Neuroblastoma may arise from problems with embryonic nerve development
Posted by lukeawylie, on 1 May 2015
Neuroblastoma is a tumour derived from the peripheral nervous system and is the most common cancer diagnosed within the first year of life. Although is a fairly rare disease, it does account for 15% of all pediatric cancer deaths. However, neuroblastoma is quite unique in that some, particularly very young, patients spontaneously regress requiring only clinical observation. Despite significant advances in the field, the underlying cause or driving mutations behind neuroblastoma have yet to be understood. Interestingly while most other cancers have significant genetic mutations underlying their disease, neuroblastoma cells on the whole appear to have their genomes largely intact, suggesting an alternative cause.
Given that the majority of neuroblastoma cases present during infancy, this implies that there is a unique window of susceptibility during development of the nervous system. During embryonic development, cells must maintain a fine balance between division, to properly generate enough cells, and the process of differentiation required for cells to perform specified functions within the embryo. Neuroblastoma arises from neural crest cells that go on to form the peripheral nervous system. Our work suggests that neuroblastoma may arise during a critical period of development because immature developing cells are incorrectly pushed towards division rather than differentiation.
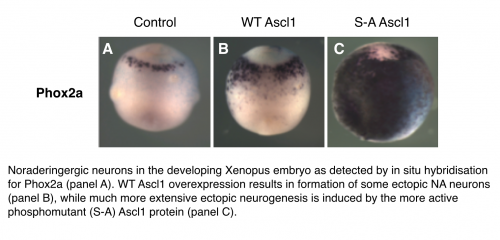
Using a widely used experimental system, tadpoles of the frog Xenopus laevis that develop outside the mother and so are almost uniquely accessible to observation and experimental manipulation of early embryonic stages, we have studied early development of peripheral nervous system. In work funded by the Neuroblastoma Society and recently published in the Journal “Disease Models and Mechanisms”, we identified a cell population of noradrinergic (NA) neurons in the developing tadpoles that are analogous to the immature nerves from which neuroblastoma is thought to derive. We then looked at genes turned on and off in these cells as the embryos progress through development and found that many genes such as Phox2a, Hand2 and Tyrosine Hydroxylase known to be expressed in NA cells in the tadpole are also found at high levels in NB. Most interestingly, we identified one key transcriptional regulator, Ascl1, that is normally only transiently expressed in normal development of NA cells but is found expressed in in both NB primary tumours and almost all the different types of NB cells we looked at. Ascl1, is a member of the basic helix-loop-helix transcription factor family, and we and others have found that is plays an important role in the switch between cell division and differentiation in a variety of neurons in the developing central and peripheral nervous systems. The fact that Ascl1 is expressed in NB cells could mean that it is aberrantly reactivated or that neuroblastoma results from a developmental stage at before Ascl1 downregulation has occurred. As well as offering a more complete understanding of the developmental stage of the neural crest from which NB derives, this also led us to explore whether the presence of Ascl1 may be of functional significance in development of neuroblastoma.
Building on our previous work in the central nervous system, we knew that the Ascl1 protein can be phosphorylated on multiple serine-proline (S)P sites and this phosphorylation limits its ability to drive neurogenesis; conversely a phosphomutant form of Ascl1 is much more effective at driving differentiation. We observed similar phospho-regulation of Ascl1 in the formation of NA neurons of the peripheral nervous system. Serine-proline sites are potential targets for cyclin-dependent kinases, as well as Map kinases, GSK3beta and other proline-directed kinases. Mechanistically, multi-site phosphorylation of Ascl1 on these sites limits its association with promoters and enhancers of downstream targets, and this prevents activation of these multiple targets that are needed for the differentiation process. Indeed, this regulation may be more wide-spread among proneural transcription factors as we have also seen similar multi-site phospho-regulation of a related proneural transcription factor Neurogenin2.
Cdk-mediated phosphorylation of Ascl1 resulting in an inhibition of its ability to drive neuronal differentiation might provide a direct link between the cellular environment in rapidly proliferating cells, and the failure to activate genes that are absolutely required for differentiation. Supporting this hypothesis, when we overexpressed Cyclin A with Cdk2 NA neuron differentiation driven by wild-type Ascl1 was inhibited, but not that driven by expression of phosphomutant Ascl1. We saw a similar phenomenon when overexpressing the N-Myc protein in the embryo; ie that N-Myc inhibits the neurogenic activity of wild-type but not phosphomutant Ascl1 It is of note that NB as whole appears to be a CDK driven disease and that the MYCN gene is very commonly amplified in poor prognosis Neuroblastoma. This suggests that in both Xenopus development and neuroblastoma cells, Ascl1 phosphorylation in response to high levels of Cdks and/or N-Myc results in suppression of NA neuronal differentiation. However, if dephosphorylation of Ascl1 is sufficient to induce NA cell differentiation in Xenopus embryos, the same may be true in neuroblastoma where we find phosphorylated Ascl1 endogenously expressed. Forcing differentiation of NB cells would be expected to confer a much more favourable prognosis than that expected when NB cells are rapidly dividing; preventing phosphorylation of Ascl1 by Cdk inhibitors, possibly in combination with other inhibitors of proline-directed kinases, is a very real potential new treatment approach for NB.
From this work, we would highlight two important conclusions: that neuroblastoma may arise during the phase in sympathetic development when Ascl1 is transiently expressed, and that Ascl1 phosphoregulation plays an important role in control of NA neuron differentiation in normal development, a regulation that may be disrupted in neuroblastoma. From a wider perspective, our findings indicate that neuroblastoma may arise from an abnormality of arrested neuronal differentiation, and so can be viewed as a disease of development. Therefore, understanding the mechanisms underlying the normal development of the peripheral nervous system and how this differs from NB cell behavior will provide critical incite into both what goes wrong during the initial events of neuroblastoma formation, and also to develop future therapies to guide neuroblastoma cells back down their normal path of differentiation.
Wylie, L., Hardwick, L., Papkovskaia, T., Thiele, C., & Philpott, A. (2015). Ascl1 phospho-status regulates neuronal differentiation in a Xenopus developmental model of neuroblastoma Disease Models & Mechanisms, 8 (5), 429-441 DOI: 10.1242/dmm.018630


 (3 votes)
(3 votes)
 – Jill and Yoan wrote about their recent paper in eLife examining how
– Jill and Yoan wrote about their recent paper in eLife examining how 
 (No Ratings Yet)
(No Ratings Yet)

 Tubular structures, such as kidney tubules or blood vessels, carry out crucial functions in organisms. Their morphogenesis requires an orchestrated sequence of cellular rearrangements, the disruption of which leads to tubule dysfunction, as observed in polycystic kidney disease. While the initiation of lumen formation is well understood, less is known about the process of lumen expansion. Using time-lapse analysis of the Ciona intestinalis notochord, a simple model of tubulogenesis in which a lumen forms between two cells connected by an apical ring of cell-cell junctions, Di Jiang and co-workers (p.
Tubular structures, such as kidney tubules or blood vessels, carry out crucial functions in organisms. Their morphogenesis requires an orchestrated sequence of cellular rearrangements, the disruption of which leads to tubule dysfunction, as observed in polycystic kidney disease. While the initiation of lumen formation is well understood, less is known about the process of lumen expansion. Using time-lapse analysis of the Ciona intestinalis notochord, a simple model of tubulogenesis in which a lumen forms between two cells connected by an apical ring of cell-cell junctions, Di Jiang and co-workers (p.  The primary cilium is an antenna-like structure present at the surface of most cells and necessary for normal development. In particular, it is required for Shh signalling, a crucial developmental pathway. However, the molecular mechanisms underlying the connection between the cilium and Shh transduction remain elusive. To address whether the ciliary localisation of Gli2, a transcriptional effector of the Shh pathway, is required for its activation, Aimin Liu and colleagues (p.
The primary cilium is an antenna-like structure present at the surface of most cells and necessary for normal development. In particular, it is required for Shh signalling, a crucial developmental pathway. However, the molecular mechanisms underlying the connection between the cilium and Shh transduction remain elusive. To address whether the ciliary localisation of Gli2, a transcriptional effector of the Shh pathway, is required for its activation, Aimin Liu and colleagues (p. The cerebellum, a posterior part of the brain crucial for motor coordination, is composed of folia – functionally distinct units that each receive specific combinations of inputs from the rest of the nervous system. Generation of folia, separated by fissures with anchoring centres at their base, requires the proliferation of granule cell progenitors (gcps) and their differentiation into granule cells (gcs). The basic pattern of folia (relative size, number) is conserved across species. To investigate how gcp behaviour regulates folium geometry, Alexandra Joyner and co-workers (p.
The cerebellum, a posterior part of the brain crucial for motor coordination, is composed of folia – functionally distinct units that each receive specific combinations of inputs from the rest of the nervous system. Generation of folia, separated by fissures with anchoring centres at their base, requires the proliferation of granule cell progenitors (gcps) and their differentiation into granule cells (gcs). The basic pattern of folia (relative size, number) is conserved across species. To investigate how gcp behaviour regulates folium geometry, Alexandra Joyner and co-workers (p.  In tissues, niche-derived signals promote stem cell self-renewal and the spatially restricted environment shields stem cells from differentiating signals, thus maintaining the stem cell pool. In an open niche environment, such as the seminiferous tubules, where both self-renewal and differentiating signals are ubiquitously distributed, it is unclear how stem cells are maintained in an undifferentiated state. In this study, Shosei Yoshida and colleagues (p.
In tissues, niche-derived signals promote stem cell self-renewal and the spatially restricted environment shields stem cells from differentiating signals, thus maintaining the stem cell pool. In an open niche environment, such as the seminiferous tubules, where both self-renewal and differentiating signals are ubiquitously distributed, it is unclear how stem cells are maintained in an undifferentiated state. In this study, Shosei Yoshida and colleagues (p. 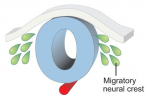 The neural crest is a uniquely vertebrate cell type and has been well studied in a number of model systems. Here, Bariga, Trainor, Bronner and Mayor discuss species-specific differences in neural crest development, urging the community to consider these differences and highlighting the need for further research in complementary systems. See the Spotlight on p.
The neural crest is a uniquely vertebrate cell type and has been well studied in a number of model systems. Here, Bariga, Trainor, Bronner and Mayor discuss species-specific differences in neural crest development, urging the community to consider these differences and highlighting the need for further research in complementary systems. See the Spotlight on p.  Sensory hair cells are mechanoreceptors of the auditory and vestibular systems and are crucial for hearing and balance. Auditory hair cells in adult mammals are unable to regenerate whereas hair cells in the chick cochlea and the zebrafish lateral line are, prompting studies into the factors that regulate hair cell development and regeneration in various species. Here, Cheng and co-workers review these studies. See the Review on p.
Sensory hair cells are mechanoreceptors of the auditory and vestibular systems and are crucial for hearing and balance. Auditory hair cells in adult mammals are unable to regenerate whereas hair cells in the chick cochlea and the zebrafish lateral line are, prompting studies into the factors that regulate hair cell development and regeneration in various species. Here, Cheng and co-workers review these studies. See the Review on p. 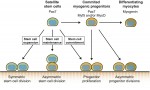 Muscle stem cells, termed satellite cells, are crucial for skeletal muscle growth and regeneration. Here, Rudnicki and colleagues review recent discoveries of the intrinsic and extrinsic factors that regulate satellite cell behaviour in regenerating and degenerating muscles. See the Review on p.
Muscle stem cells, termed satellite cells, are crucial for skeletal muscle growth and regeneration. Here, Rudnicki and colleagues review recent discoveries of the intrinsic and extrinsic factors that regulate satellite cell behaviour in regenerating and degenerating muscles. See the Review on p.