Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9
Posted by Shashank Gandhi, on 31 October 2014
Researchers have always been interested in tissue-specific loss of function to probe the role of specific genes in embryonic development, cell physiology and disease conditions. Migration of lateral plate primordial germ cells in zebrafish, border cell migration during oogenesis in drosophila, interaction of T-cells with their target, and numerous other cases have continued to give us insights about how cells behave differently depending on their position and function in the animal. Until recently, genome-editing approaches were limited to non-specific mutations, induced by chemical or transposable elements. Homologous recombination for precise genome editing has only been achieved in mouse embryonic stem cells. Our lab studies the embryonic development and gene expression in Ciona intestinalis, a hermaphroditic tunicate. Phylogenetic studies have shown that tunicates are the closest living relatives of vertebrates. The Ciona larva possesses the basic body plan of chordates. A small sequenced genome, low cell number (~2600 at the larval stage), well-defined cell lineages, and easy methods for transient transfection assays are some of the experimental advantages offered by this model chordate. Previous studies have reported high rate of mutagenesis and tissue-specific gene knockouts in Ciona intestinalis using TALENs and ZFNs, but the application of these methods has been limited because of expensive and tedious cloning procedures.
An adult Ciona intestinalis (Copyright: John Rundle)
Significant technical breakthroughs have been achieved in the field of genome engineering by harnessing the CRISPR (Clustered, Regularly Interspaced, Short Palindromic Repeats)/Cas (CRISPR Associated) system, an important part of the prokaryotic immune response. In this system, a short sequence, known as protospacer guides the endonuclease Cas9 to specific sites on the genome where it induces double stranded breaks. A broad range of applications, like genome editing, knock-in of exogenous DNA fragments, and regulation of transcription of endogenous genes, in a wide variety of model organisms, have demonstrated the versatility of this system. In our Development paper, we have established the CRISPR/Cas9 system for tissue specific genome editing in Ciona embryos using a simple electroporation-based transfection technique to induce site-specific double stranded breaks (DSBs) in the Ciona genome. High mutation efficiencies observed in our study are in sync with the results reported by Sasakura et al (2014). The main highlights of our paper include tissue-specific mutagenesis obtained using CRISPR/Cas system, and the phenotypic effects observed as a result of double stranded breaks in Ciona embryos.
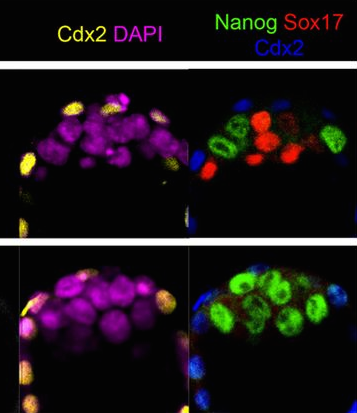
We have demonstrated that electroporation-based usage of the CRISPR/Cas system is sufficient to disrupt the activity of Ebf (previously known as COE), the sole C. intestinalis homolog of vertebrate EBF1, -2, -3 and -4. Ebf has been reported to play critical roles in both the ectoderm-derived motor neurons and in the cardiogenic mesoderm-derived Atrial Siphon Muscles (ASM), for their specification at the expense of heart development in Ciona. Ebf was thought to specifically up-regulate Islet expression in both the motor neurons and ASM precursors. To drive the expression of Cas9 in the ectoderm, we used the upstream cis-regulatory sequences of the C. intestinalis Sox1/2/3 gene. We observed a down-regulation of Islet reporter activity in motor ganglion neurons and ASM precursors, validating that Ebf is required for the specification and differentiation of motor ganglion neurons and pharyngeal muscles in Ciona intestinalis. Expression of a CRISPR/Cas9-resistant form of Ebf (Ebf>Ebfm774) was sufficient to rescue Islet expression, suggesting that the observed phenotypes were specific consequences of Cas9-mediated mutagenesis of Ebf gene. Moreover, when Ebf was targeted in the cardiogenic mesoderm by Mesp enhancer driven Cas9, Islet reporter expression was lost in the ASM but unaffected in motor ganglion neurons, confirming the tissue-specificity of CRISPR/Cas9 system in Ciona.
Spatial variation in expression of exogenous DNA associated with electroporation, known as mosaicism, is commonly associated with transient transfection assays. It has been shown to result in electroporated embryos containing both transfected and non-transfected cells. The latter would not experience CRISPR/Cas9-mediated mutagenesis, thus potentially leading us to underestimate mutagenesis efficacy. To address this problem, we used a simple cell-enrichment technique called MACS (Magnetic Activated Cell Sorting). Transfected cells expressed the guide RNA vector and Cas9 alongside the membrane-bound reporter hCD4::mCherry, which served for affinity purification using anti-hCD4 antibodies conjugated to magnetic nano-beads. Following one-step incubation with the beads, the sample was applied to a MACS column. The magnetically labeled cells were retained within the column, and were later eluted, whereas the unlabeled cells passed through. The sorted cells exhibited 66.2% mutagenesis when compared to unsorted cells from the same pool of embryos, which had 45.1% mutagenesis. Hence, a significant increase in the estimation of mutagenesis rates was observed using this simple technique. Furthermore, studying the temporal dynamics of Ci-EF1α promoter revealed the onset of its activity at 5 hours post fertilization (hpf) at the earliest, suggesting that the microinjection of mRNA transcripts into Ciona embryos might be necessary in order to target genes that are expressed before the gastrula stage.
Recently, the Sasakura lab at the University of Tsukuba published a paper demonstrating CRISPR/Cas9-mediated knockout of the Ci-Hox3 and Ci-Hox5 genes in Ciona intestinalis. They compared microinjection of in-vitro transcribed single guide RNA (sgRNA) and Cas9 mRNA transcripts with an electroporation-based approach to induce double stranded breaks in the Ciona genome. They observed an increase in the rate of mutagenesis when more RNA transcripts were microinjected into the embryos, although, the authors have not addressed questions related to tissue specificity and the efficacy of CRISPR/Cas9 induced mutations to cause specific phenotypes. One of the key findings of their paper was the high sensitivity of CRISPR/Cas9 mediated mutagenesis to the sequence of sgRNA vectors and mRNA transcripts. They observed a complete loss in mutation frequency with even a single nucleotide difference in the protospacer sequence, albeit reports of significant off-target effects of Cas9 in various model organisms. The authors of the paper have used instability of the Cas9-sgRNA complex formed on the genomic DNA as an explanation for this observation. However, we believe that this observation could be a result of using an unstable sgRNA backbone, an issue that has been addressed in our article. Two modifications, namely Flip (F) and Elongation (E), have been suggested in the guide RNA backbone to increase its in-vivo transcription efficiency, at the same time, promoting its ability to form a stable complex with Cas9. Using a modified backbone, we were able to achieve mutations in the 5’ flanking regions of Foxf and Hand-related, suggesting that CRISPR/Cas9 could be used for targeted mutagenesis in a wide variety of loci in the Ciona genome.
In conclusion, both papers report the successful application of the CRISPR/Cas system for effective genome editing in Ciona intestinalis. An expression vector-mediated electroporation method enables validation and extensive screening of sgRNA vectors, which is essential in the absence of a well-defined designing criterion to obtain high mutational activity. That being said, a microinjection-based approach might be necessary to construct loss-of-function sgRNA library to target genes that are expressed at an early stage during embryogenesis. Either ways, the CRISPR/Cas system has the potential to serve for the rapid scaling of genome editing in this model chordate.
Reference:
Stolfi, A., Gandhi, S., Salek, F., & Christiaen, L. (2014). Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9 Development, 141 (21), 4115-4120 DOI: 10.1242/dev.114488



 (6 votes)
(6 votes)

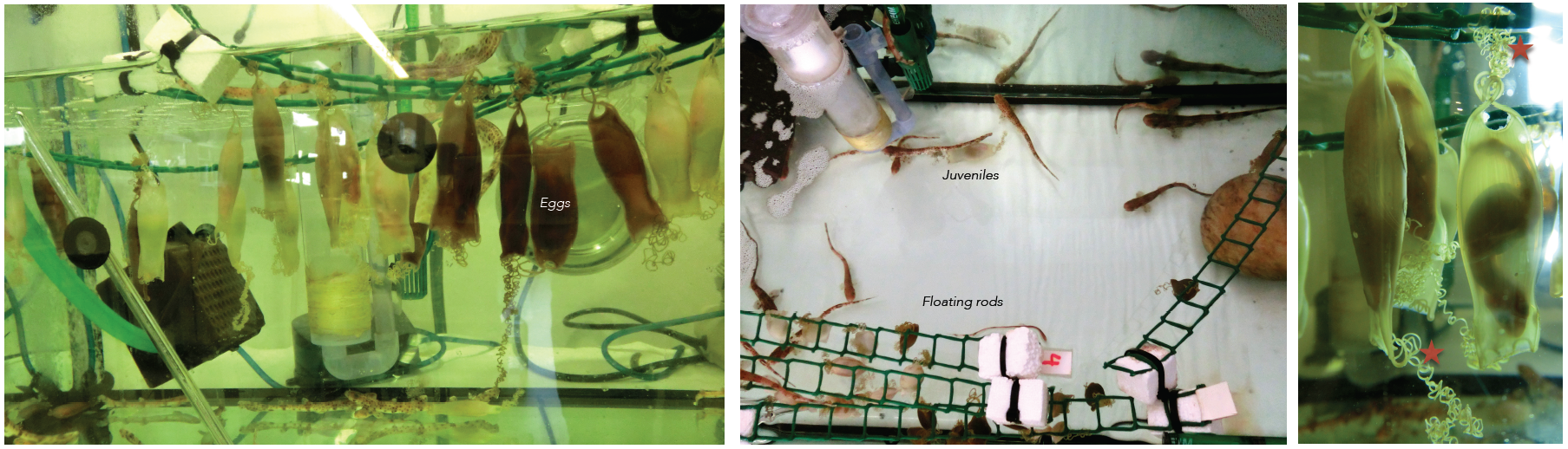
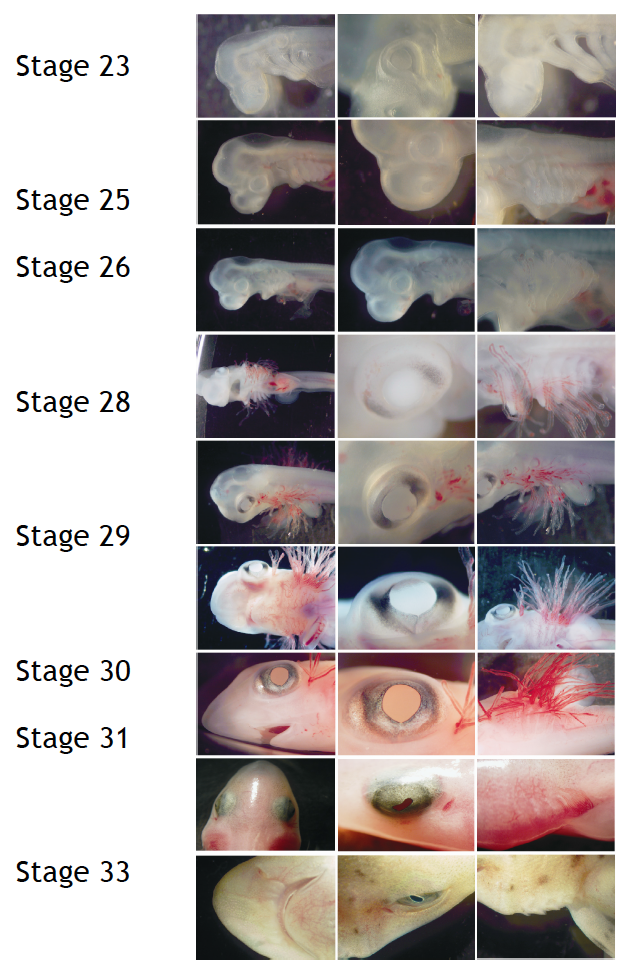

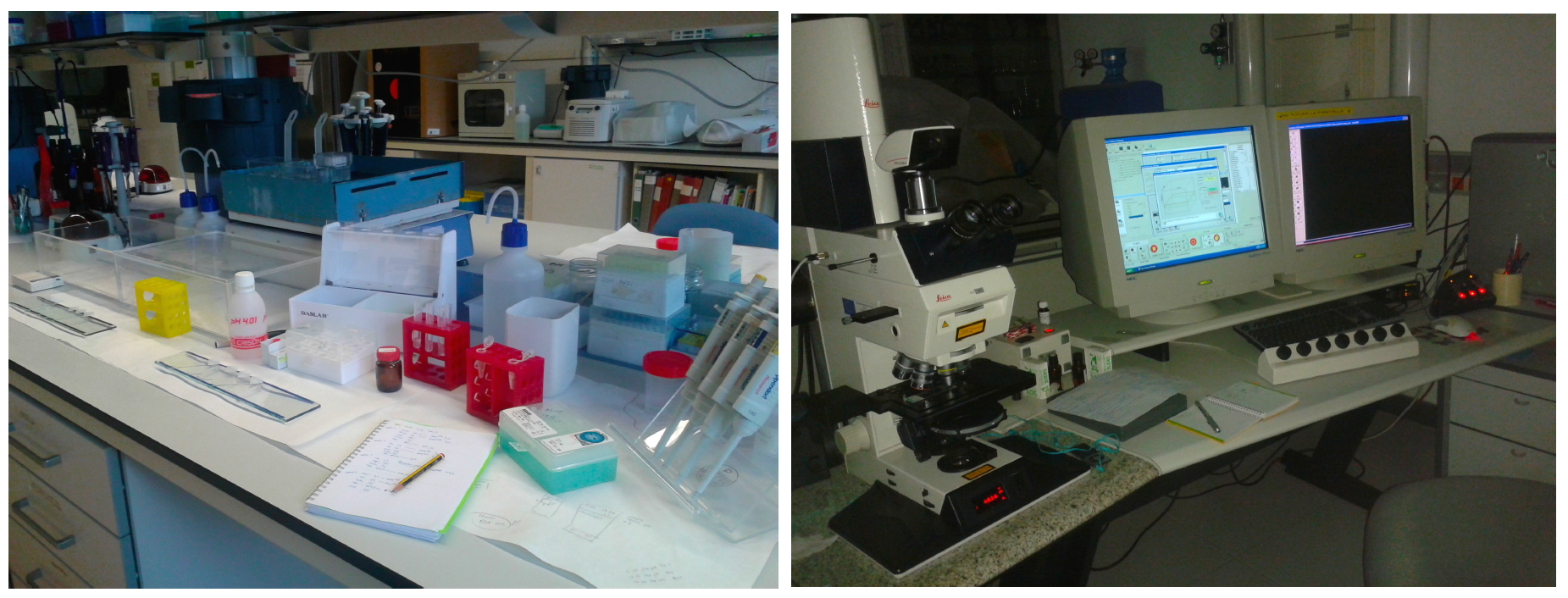
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series 

 (No Ratings Yet)
(No Ratings Yet)


 The widely accepted model of angiogenic sprouting proposes that a single cell – the tip cell – is found at the leading edge of vessel sprouts. Now, Victoria Bautch and colleagues describe an alternative blood vessel topology in which multiple endothelial cells (ECs) constitute the tip of sprouting blood vessels and polarize to promote lumen formation (p.
The widely accepted model of angiogenic sprouting proposes that a single cell – the tip cell – is found at the leading edge of vessel sprouts. Now, Victoria Bautch and colleagues describe an alternative blood vessel topology in which multiple endothelial cells (ECs) constitute the tip of sprouting blood vessels and polarize to promote lumen formation (p.  Development of the Drosophila tracheal system – the fly respiratory network – is known to require the chitin deacetylase serpentine (Serp), which is expressed in tracheal epithelial cells. Here, Shigeo Hayashi and co-workers show that Serp derived from the Drosophila fat body, which is functionally equivalent to the mammalian liver, is transported to the tracheal lumen and is required for tube morphogenesis (p.
Development of the Drosophila tracheal system – the fly respiratory network – is known to require the chitin deacetylase serpentine (Serp), which is expressed in tracheal epithelial cells. Here, Shigeo Hayashi and co-workers show that Serp derived from the Drosophila fat body, which is functionally equivalent to the mammalian liver, is transported to the tracheal lumen and is required for tube morphogenesis (p.  The cerebral hemispheres are connected by the largest axonal tract in the mammalian brain: the corpus callosum (CC). During CC development, axons from one hemisphere navigate across the midline, channelled along their way by guidepost cells that secrete guidance cues, to reach specific targets on the other hemisphere. Neurofibromatosis type 2 (Nf2/Merlin) mouse mutants show a complete absence of the CC but how Nf2, a signalling protein involved in various cellular processes and signalling pathways, controls axonal pathfinding is currently unclear. Using Nf2-conditional knockout mouse models, Xinwei Cao and colleagues show that, surprisingly, Nf2 is not required in callosal neurons or their progenitors but is required in midline neural progenitors that generate guidepost cells (see p.
The cerebral hemispheres are connected by the largest axonal tract in the mammalian brain: the corpus callosum (CC). During CC development, axons from one hemisphere navigate across the midline, channelled along their way by guidepost cells that secrete guidance cues, to reach specific targets on the other hemisphere. Neurofibromatosis type 2 (Nf2/Merlin) mouse mutants show a complete absence of the CC but how Nf2, a signalling protein involved in various cellular processes and signalling pathways, controls axonal pathfinding is currently unclear. Using Nf2-conditional knockout mouse models, Xinwei Cao and colleagues show that, surprisingly, Nf2 is not required in callosal neurons or their progenitors but is required in midline neural progenitors that generate guidepost cells (see p.  During a heart beat, an electrical impulse leads to the contraction of the upper part of the heart: the atria. The electrical impulse slows down through the atrioventricular junction (AVJ) before resuming rapid propagation to induce contraction of the lower part of the heart: the ventricles. This contraction delay, combined with the presence of cardiac valves, is crucial for unidirectional blood flow in the heart and is altered in various heart diseases. How is this slow conducting property established and restricted to the AVJ? On p.
During a heart beat, an electrical impulse leads to the contraction of the upper part of the heart: the atria. The electrical impulse slows down through the atrioventricular junction (AVJ) before resuming rapid propagation to induce contraction of the lower part of the heart: the ventricles. This contraction delay, combined with the presence of cardiac valves, is crucial for unidirectional blood flow in the heart and is altered in various heart diseases. How is this slow conducting property established and restricted to the AVJ? On p. The cerebellum is a pre-eminent model for the study of neurogenesis and circuit assembly.
The cerebellum is a pre-eminent model for the study of neurogenesis and circuit assembly.  Recent advances in the targeted modification of eukaryotic genomes have unlocked a new era of genome engineering. From pioneering work using zinc-finger nucleases, to the advent of TALEN and CRISPR/Cas9 systems, we now possess the ability to analyze developmental processes using sophisticated genetic tools. Here, Stephen Ekker and colleagues summarize the common approaches and applications of these still-evolving tools. See the Review on p.
Recent advances in the targeted modification of eukaryotic genomes have unlocked a new era of genome engineering. From pioneering work using zinc-finger nucleases, to the advent of TALEN and CRISPR/Cas9 systems, we now possess the ability to analyze developmental processes using sophisticated genetic tools. Here, Stephen Ekker and colleagues summarize the common approaches and applications of these still-evolving tools. See the Review on p.