Outreach Activity – Smartphone to Microscope Conversion
Posted by Alexis Webb, on 6 May 2014
Development is a fascinating process that few people have a chance to see, let alone photograph! We recently participated with other scientists from the Crick Institute at a Science Museum Lates in London in February. For our activity, we built these inexpensive platforms that convert a user’s smartphone into a microscope screen.
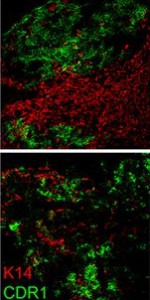
We provided zebrafish embryos at different developmental stages for visitors to visualize and photograph. We showed visitors how to use the simple platforms that include a lens from a laser pointer for magnification. Once visitors had their phones lined up on the platform, they were able to view individual cells in early stage embryos or structures like the eye, brain, and heart in older specimens. We described the process of development from cell divisions to cell movements, gastrulation and segmentation. We found that the best practice was to mount embryos in sealed, agarose-coated petri dishes. This kept the embryo medium and zebrafish contained and the dishes were easy for visitors to manipulate. We also found that focal plane could be slightly different for various phones; it was helpful for visitors to remove phones from their cases.
Photo credit: Thomas S. G. Farnetti/Wellcome Images
Visitors were very excited to be able to take images like these with their smartphones. The event produced several shares on social media sites like Twitter and Instagram of pictures of zebrafish.
Photo credit: Alexis Webb
These microscope platforms are affordable and portable, making them suitable for demonstrations in schools and classrooms. Because they don’t require any power, they can be used outdoors or in areas that do not have electrical outlets. Imaging other types of live specimens is also possible. We hope that other researchers interested in showing off their model organism will consider using this type of low-tech, but high reward activity!
 This post is part of a series on science outreach. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on science outreach. You can read the introduction to the series here and read other posts in this series here.




 (2 votes)
(2 votes) Adult stem cells play crucial roles in tissue homeostasis, giving rise to both new stem cells and differentiating daughter cells. The generation of these two cell types often involves the asymmetric distribution of cell fate determinants, but how these factors are partitioned asymmetrically has been unclear. Now (p.
Adult stem cells play crucial roles in tissue homeostasis, giving rise to both new stem cells and differentiating daughter cells. The generation of these two cell types often involves the asymmetric distribution of cell fate determinants, but how these factors are partitioned asymmetrically has been unclear. Now (p.  It is widely accepted that, in amniotes, WNTs secreted by the dorsal neural tube form a concentration gradient that regulates somite patterning and myotome organisation. Here, Olivier Serralbo and Christophe Marcelle challenge this assumption and uncover a novel mode of long-range WNT signalling in which WNTs are delivered to their target sites by migratory neural crest cells (p.
It is widely accepted that, in amniotes, WNTs secreted by the dorsal neural tube form a concentration gradient that regulates somite patterning and myotome organisation. Here, Olivier Serralbo and Christophe Marcelle challenge this assumption and uncover a novel mode of long-range WNT signalling in which WNTs are delivered to their target sites by migratory neural crest cells (p.  Hox genes play a crucial role in assigning cellular identities along the anterior-posterior axis of animal bodies. Hox gene expression can be regulated via transcriptional mechanisms and recent studies have also uncovered a regulatory role for Hox RNA processing, yet the mechanisms underlying this regulation remain unknown. Now, Claudio Alonso and colleagues identify the neural RNA-binding protein ELAV as a key regulator of Hox RNA processing in the Drosophila embryonic central nervous system (p.
Hox genes play a crucial role in assigning cellular identities along the anterior-posterior axis of animal bodies. Hox gene expression can be regulated via transcriptional mechanisms and recent studies have also uncovered a regulatory role for Hox RNA processing, yet the mechanisms underlying this regulation remain unknown. Now, Claudio Alonso and colleagues identify the neural RNA-binding protein ELAV as a key regulator of Hox RNA processing in the Drosophila embryonic central nervous system (p.  A locus in mice known as strain-specific modifier 1 (Ssm1) has previously been shown to be responsible for the strain-dependent methylation of E. coli gpt-containing transgenic sequences. Now, Ursula Storb and co-workers identify the Ssm1b gene that underlies this phenotype and characterise its expression in early mouse embryos (p.
A locus in mice known as strain-specific modifier 1 (Ssm1) has previously been shown to be responsible for the strain-dependent methylation of E. coli gpt-containing transgenic sequences. Now, Ursula Storb and co-workers identify the Ssm1b gene that underlies this phenotype and characterise its expression in early mouse embryos (p.  Adult neurogenesis has been implicated in physiological brain function, and failing or altered neurogenesis has been associated with a number of neuropsychiatric diseases. Simon Braun and Sebastian Jessberger provide an overview of the mechanisms governing the neurogenic process in the adult brain and describe how new neurons may contribute to brain function in health and disease. See the Development at a Glance poster article on p.
Adult neurogenesis has been implicated in physiological brain function, and failing or altered neurogenesis has been associated with a number of neuropsychiatric diseases. Simon Braun and Sebastian Jessberger provide an overview of the mechanisms governing the neurogenic process in the adult brain and describe how new neurons may contribute to brain function in health and disease. See the Development at a Glance poster article on p.  Apical constriction is a cell shape change that promotes tissue remodelling in a variety of contexts. Martin and Goldstein review the cellular machinery required for apical constriction and discuss how it can be tunedto regulate apical constriction in diverse cellular contexts. See the Review article on p.
Apical constriction is a cell shape change that promotes tissue remodelling in a variety of contexts. Martin and Goldstein review the cellular machinery required for apical constriction and discuss how it can be tunedto regulate apical constriction in diverse cellular contexts. See the Review article on p.  Cell migration is a fundamental process that occurs during embryo development. Here, Concha and colleagues review the guidance principles of in vitro cell locomotion and examine how they apply to examples of directed cell migration observed in vivo during development. See the Review on p.
Cell migration is a fundamental process that occurs during embryo development. Here, Concha and colleagues review the guidance principles of in vitro cell locomotion and examine how they apply to examples of directed cell migration observed in vivo during development. See the Review on p.  (No Ratings Yet)
(No Ratings Yet)