February in preprints
Posted by the Node, on 9 March 2022
Welcome to our monthly trawl for developmental and stem cell biology (and related) preprints.
The preprints this month are hosted on bioRxiv and arXiv– use these links to get to the section you want.
- Patterning & signalling
- Morphogenesis & mechanics
- Genes & genomes
- Stem cells, regeneration & disease modelling
- Plant development
- Evo-devo
Developmental biology
| Patterning & signalling
Atypical MAPK regulates translocation of GATA transcription factor in response to chemoattractant stimulation
Jeffrey Hadwiger, Huaqing Cai, Ramee Aranda, Saher Fatima
Comparisons of cell proliferation and cell death across life histories in the hemichordate Schizocardium californicum
Paul Bump, Margarita Khariton, Clover Stubbert, Nicole E Moyen, Jia Yan, Bo Wang, Christopher J Lowe
Ecdysone coordinates plastic growth with robust pattern in the developing wing
Andre Nogueira Alves, Marisa Mateus Oliveira, Takashi Koyama, Alexander Shingleton, Christen Mirth
The Drosophila melanogaster enzyme glycerol-3-phosphate dehydrogenase 1 is required for oogenesis, embryonic development, and amino acid homeostasis
Madhulika Rai, Sarah M. Carter, Shefali A. Shefali, Nader H. Mahmoudzadeh, Robert Pepin, Jason M. Tennessen
A gradient border model for cell fate decisions at the neural plate border
Alexandre Thiery, Ailin Leticia Buzzi, Eva Hamrud, Chris Cheshire, Nicholas Luscombe, James Briscoe, Andrea Streit
Canonical Wnt signaling promotes formation of somatic permeability barrier for proper germ cell differentiation
Ting-An Chen, Kun-Yang Lin, Shun-Min Yang, Chen-Yuan Tseng, Yu-Ting Wang, Chi-Hung Lin, Lichao Luo, Yu Cai, Hwei-Jan Hsu
Prolactin-induced AMPK stabilizes alveologenesis and lactogenesis through regulation of STAT5 signaling
Shyam Lal Jinagal, Pragati Shekhar, Kailash Chandra, Srinivas Abhishek Mutnuru, Narendrakumar Ramanan, Marc Foretz, Benoit Viollet, Ramray Bhat, Annapoorni Rangarajan
The calcium channel Orai1 is required for osteoblast development: studies in a chimeric mouse with variable in vivo Runx-cre deletion of Orai-1
Lisa J Robinson, Jonathan Soboloff, Irina L Tourkova, Quitterie C Larrouture, Dionysios J Papachristou, Scott Gross, Robert Hooper, Elsie Samakai, Paul F Worley, Jan Tuckermann, Michelle R Witt, Harry C Blair
A timer gene network is spatially regulated by the terminal system in the Drosophila embryo
Erik Clark, Margherita Battistara, Matthew A. Benton
Completion of neural crest cell production and emigration is regulated by retinoic acid-dependent inhibition of BMP signaling
Dina Rekler, Chaya Kalcheim
ERK1/2 is an ancestral organising signal in spiral cleavage
Océane Seudre, Allan M. Carrillo-Baltodano, Yan Liang, José M. Martín-Durán
Timely Schwann cell division during migration drives peripheral myelination in vivo via Laminin/cAMP pathway
Aya Mikdache, Marie-José Boueid, Emilie Lesport, Brigitte Delespierre, Julien Loisel-Duwattez, Cindy Degerny, Marcel Tawk
Antennapedia and optix regulate metallic silver wing scale development and cell shape in Bicyclus anynana butterflies
Anupama Prakash, Cédric Finet, Tirtha Das Banerjee, Vinodkumar Saranathan, Antónia Monteiro
Visceral mesoderm signaling regulates assembly position and function of the Drosophila testis niche
Lauren Anllo, Stephen DiNardo
Synthetic reconstruction of the hunchback promoter specifies the role of Bicoid, Zelda and Hunchback in the dynamics of its transcription
Goncalo Fernandes, Huy Tran, Maxime Andrieu, Youssoupha Diaw, Carmina Perez Romero, Cécile Fradin, Mathieu Coppey, Aleksandra M. Walczak, Nathalie Dostatni
unc-37/Groucho and lsy-22/AES repress Wnt target genes in C. elegans asymmetric cell divisions
Kimberly N. Bekas, Bryan T. Phillips
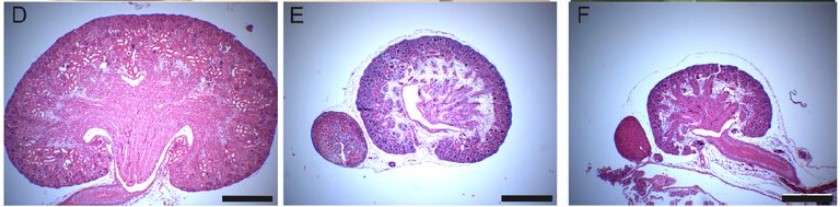
FGF8 induces chemokinesis and regulates condensation of mouse nephron progenitor cells
Abhishek Sharma, Marco Meer, Arvydas Dapkunas, Anneliis Ihermann-Hella, Satu Kuure, Seppo Vainio, Dagmar Iber, Florence Naillat
Distinct mechanisms of germ cell factor regulation for an inductive germ cell fate
Stephany Foster, Nathalie Oulhen, Tara Fresques, Hossam Zaki, Gary Wessel
Maternal Wnt11b regulates cortical rotation during Xenopus axis formation: analysis of maternal-effect wnt11b mutants
Douglas W. Houston, Karen L. Elliott, Kelsey Coppenrath, Marcin Wlizla, Marko E. Horb
Impact of cell size on morphogen gradient precision
Jan A. Adelmann, Roman Vetter, Dagmar Iber
Trunk neural crest migratory position and asymmetric division predict terminal differentiation
Zain Alhashem, Karen Camargo-Sosa, Robert N Kelsh, Claudia Linker
Mechano-signaling feedback underlies precise inner hair cell patterning in the organ of Corti
Roie Cohen, Shahar Taiber, Olga Loza, Shahar Kasirer, Shiran Woland, David Sprinzak
Sonic Hedgehog is not a limb morphogen but acts as a trigger to specify all digits
Jianjian Zhu, Rashmi Patel, Anna Trofka, Brian D. Harfe, Susan Mackem
Photoreceptors generate neuronal diversity in their target field through a Hedgehog morphogen gradient in Drosophila
Matthew P. Bostock, Vilaiwan M. Fernandes
Mural norrin/β-catenin signaling regulates Lama2 expression to promote neurovascular unit assembly
Saptarshi Biswas, Sanjid Shahriar, Nicholas P. Giangreco, Panos Arvanitis, Markus Winkler, Nicholas P. Tatonetti, William J. Brunken, Tyler Cutforth, Dritan Agalliu
Atypical MAPK regulates translocation of GATA transcription factor in response to chemoattractant stimulation
Jeffrey A. Hadwiger, Huaqing Cai, Ramee G. Aranda, Saher Fatima
Hypoxia promotes osteogenesis via regulation of the mito-nuclear communication
Andromachi Pouikli, Monika Maleszewska, Swati Parekh, Chrysa Nikopoulou, Juan-Jose Bonfiglio, Constantine Mylonas, Tonantzi Sandoval, Anna-Lena Schumacher, Yvonne Hinze, Ivan Matic, Peter Tessarz
Human pluripotent stem cell-derived cardiomyocytes align under cyclic strain when guided by cardiac fibroblasts
Dylan Mostert, Bart Groenen, Leda Klouda, Robert Passier, Marie-Jose Goumans, Nicholas A. Kurniawan, Carlijn V.C Bouten
AIMP1-derived peptide secreted from hair follicle stem cells activates dermal papilla cells to promote hair growth
YounHa Kim, Ho Lee, Doyeun Kim, Soon Sun Bak, Ina Yoon, Ralf Paus, Seongmin Cho, Seung Jae Jeong, Yoon Jeon, Min Chul Park, Ji Won Oh, Jung Min Park, Sang Bum Kim, Young Kwan Sung, Sunghoon Kim
Live-Cell Imaging in Human Colonic Monolayers Reveals Erk Waves Limit the Stem Cell Compartment to Maintain Epithelial Homeostasis
Kelvin W Pond, Olga Alkhimenok, Jayati Chakrabarti, Yana Zavros, Curtis A Thorne, Andrew L Paek
The Role of Notch Signaling in Endometrial Mesenchymal Stromal/Stem-like Cells Maintenance
Sisi Zhang, Rachel W.S. Chan, Ernest H.Y. Ng, William S.B. Yeung
G2 stem cells orchestrate time-directed, long-range coordination of calcium signaling during skin epidermal regeneration
Jessica L Moore, Feng Gao, Catherine Matte-Martone, Shuangshuang Du, Elizabeth Lathrop, Smirthy Ganesan, Lin Shao, Dhananjay Bhaskar, Andy Cox, Caroline Hendry, Bastian Rieck, Smita Krishnaswamy, Valentina Greco
The Growth Hormone Releasing Hormone Signaling Pathway Governs Cardiomyocyte differentiation in human iPS cells
Amarylis C.B.A. Wanschel, Konstantinos E. Hatzistergos, Alessandro G. Salerno, Jeffim N. Kuznetsov, Stefan Kurtenbach, Daniel A. Rodriguez, Krystalenia Valasaki, Wayne Balkan, Derek Dykxhoorn, Andrew V. Schally, Joshua M. Hare
| Morphogenesis & mechanics
Contribution of the Wolffian duct mesenchyme to the formation of the female reproductive tract
Fei Zhao, Sara A Grimm, Shuai Jia, Humphrey Hung-Chang Yao
Early zygotic gene product Dunk interacts with anillin to regulate Myosin II during Drosophila cleavage
Jiayang Chen, Bing He
Oligodendrocyte origin and development in the zebrafish visual system
Adrián Santos-Ledo, Cristina Montes-Perez, Laura DeOliveira-Mello, Rosario Arévalo, Almudena Velasco
Optogenetic dissection of actomyosin-dependent mechanics underlying tissue fluidity
R. Marisol Herrera-Perez, Christian Cupo, Cole Allan, Alicia B. Dagle, Karen E. Kasza
Compaction of Drosophila histoblasts in a crowded epidermis is driven by buckling of their apical junctions
Annafrancesca Rigato, Huicheng Meng, Faris Abouakil, Loïc LeGoff
The Requirement of Ubiquitin C-Terminal Hydrolase L1 (UCHL1) in Mouse Ovarian Development and Fertility
Morgan F. Woodman, Meghan C.H. Ozcan, Megan A. Gura, Payton De La Cruz, Alexis K. Gadson, Kathryn J. Grive
A Yap-dependent transcriptional program directs cell migration for embryo axis assembly
Ana Sousa-Ortega, Javier Vazquez-Marin, Estefanía Sanabria-Reinoso, Rocío Polvillo, Alejandro Campoy-López, Lorena Buono, Felix Loosli, María Almuedo-Castillo, Juan R. Martinez-Morales
Changes in body shape implicate cuticle stretch in C. elegans growth control
Joy Nyaanga, Christina Goss, Gaotian Zhang, Hannah N. Ahmed, Elliot J. Andersen, Isabella R. Miller, Justine K. Rozenich, Iris L. Swarthout, Jordan A. Vaughn, Niall M. Mangan, Sasha Shirman, Erik C. Andersen
Myopia alters the structural organization of the retinal astrocyte template, associated vasculature and ganglion layer thickness
Carol Lin, Abduqodir Toychiev, Nefeli Slavi, Reynolds Ablordeppey, Miduturu Srinivas, Alexandra Benavente-Perez
Comparisons of cell proliferation and cell death across life histories in the hemichordate Schizocardium californicum
Paul Bump, Margarita Khariton, Clover Stubbert, Nicole E. Moyen, Jia Yan, Bo Wang, Christopher J. Lowe
Loading-Induced Bone Formation is Mediated by Wnt1 Induction in Osteoblast-Lineage Cells
Lisa Y. Lawson, Nicole Migotsky, Christopher J. Chermside-Scabbo, John T. Shuster, Roberto Civitelli, Matthew J. Silva
Mechanical feedback controls the emergence of dynamical memory in growing tissue monolayers
Sumit Sinha, Xin Li, Rajsekhar Das, D. Thirumalai
Force sensing on cells and tissues by atomic force microscopy
Hatice Holuigue, Ewelina Lorenc, Matteo Chighizola, Carsten Schulte, Luca Varinelli, Marcello Deraco, Marcello Guaglio, Manuela Gariboldi, Alessandro Podestà
Tension at intercellular junctions is necessary for accurate orientation of cell division in the epithelium plane
Ana Lisica, Jonathan Fouchard, Manasi Kelkar, Tom P. J. Wyatt, Julia Duque, Anne-Betty Ndiaye, Alessandra Bonfanti, Buzz Baum, Alexandre J. Kabla, Guillaume T. Charras
| Genes & genomes
Sperm Histone H3 Lysine 4 tri-methylation serves as a metabolic sensor of paternal obesity and is associated with the inheritance of metabolic dysfunction
Anne-Sophie Pepin, Christine Lafleur, Romain Lambrot, Vanessa Dumeaux, Sarah Kimmins
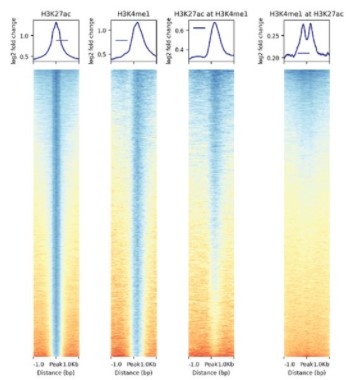
Identification of enhancer-like elements defines regulatory networks active in planarian adult stem cells
Jakke Neiro, Divya Sridhar, Anish Dattani, Aziz Aboobaker
Context-Dependent Enhancer Function Revealed by Targeted Inter-TAD Relocation
Christopher Chase Bolt, Lucille Lopez-Delisle, Aurélie Hintermann, Bénédicte Mascrez, Antonella Rauseo, Guillaume Andrey, Denis Duboule
Prediction of sex-determination mechanisms in avian primordial germ cells using RNA-seq analysis
Kennosuke Ichikawa, Yoshiaki Nakamura, Hidemasa Bono, Ryo Ezaki, Mei Matsuzaki, Hiroyuki Horiuchi
Zebrafish Neuromesodermal Progenitors Undergo a Critical State Transition in vivo
Kane Toh, Dillan Saunders, Berta Verd, Benjamin Steventon
Deconvolution of the epigenetic age discloses distinct inter-personal variability in epigenetic aging patterns
Tamar Shahal, Elad Segev, Thomas Konstantinovsky, Yonit Marcus, Gabi Shefer, Metsada Pasmanik-Chor, Assaf Buch, Yuval Ebenstein, Paul Zimmet, Naftali Stern
Estrogen suppresses DMRT1 expression during ovarian development in the chicken
Debiao Zhao, Long Liu, Sunil Nandi, Jason Ioannidis, Xiurong Yang, Daoqing Gong, Mike J. McGrew, Michael Clinton
Chromatin state transition underlies the temporal changes in gene expression during cardiomyocyte maturation
Chia-Yeh Lin, Yao-Ming Chang, Hsin-Yi Tseng, Yen-Ling Shih, Hsiao-Hui Yeh, You-Rou Liao, Chia-Ling Hsu, Chien-Chang Chen, Yu-Ting Yan, Cheng-Fu Kao
Integrated epigenome and transcriptome analysis of normal and arrested meiotic initiation during mouse spermatogenesis
Xiaoyu Zhang, Sumedha Gunewardena, Ning Wang
Contribution of the Wolffian duct mesenchyme to the formation of the female reproductive tract
Fei Zhao, Sara A Grimm, Shua Jia, Humphrey Hung-Chang Yao
Spatially resolved epigenomic profiling of single cells in complex tissues
Tian Lu, Cheen Euong Ang, Xiaowei Zhuang
The high-throughput perturbation of long non-coding RNA reveals functional features in stem cells and across cell-types
Chi Wai Yip, Chung-Chau Hon, Kayoko Yasuzawa, Divya M. Sivaraman, Jordan A. Ramilowski, Youtaro Shibayama, Saumya Agrawal, Anika V. Prabhu, Callum Parr, Jessica Severin, Yan Jun Lan, Josée Dostie, Hiromi Nishiyori-Sueki, Michihira Tagami, Masayoshi Itoh, Fernando López-Redondo, Tsukasa Kouno, Jen-Chien Chang, Joachim Luginbühl, Masaki Kato, Mitsuyoshi Murata, Wing Hin Yip, Xufeng Shu, Imad Abugessaisa, Akira Hasegawa, Harukazu Suzuki, Ken Yagi, Takeya Kasukawa, Michiel de Hoon, Piero Carninci, Jay W. Shin
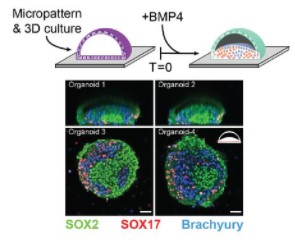
Integrated single-cell sequencing reveals principles of epigenetic regulation of human gastrulation and germ cell development in a 3D organoid model
Alex Chialastri, Eyal Karzbrun, Aimal H. Khankhel, Monte J. Radeke, Sebastian J. Streichan, Siddharth S. Dey
Genome-wide profiling of histone H3K4me3 and H3K27me3 modifications in individual blastocysts by CUT&Tag without a solid support (NON-TiE-UP CUT&Tag)
Kazuki Susami, Shuntaro Ikeda, Yoichiro Hoshino, Shinnosuke Honda, Naojiro Minami
Mutations in coral soma and sperm imply lifelong stem cell renewal and cell lineage selection
Elora H. López-Nandam, Rebecca Albright, Erik A. Hanson, Elizabeth A. Sheets, Stephen R. Palumbi
| Stem cells, regeneration & disease modelling
A unique mineralizing pool of Gli1+ stem cells builds the tendon enthesis and demonstrates therapeutic potential
Fei Fang, Yang Xiao, Elazar Zelzer, Kam W Leong, Stavros Thomopoulos
Clonal behaviour of myogenic precursor cells throughout the vertebrate lifespan.
Simon M Hughes, Roberta C Escaleira, Kees Wanders, Jana Koth, David G Wilkinson, Qiling Xu
Dynamic regulation and requirement for ribosomal RNA transcription during mammalian development
Karla T. Falcon, Kristin E.N. Watt, Soma Dash, Ruonan Zhao, Daisuke Sakai, Emma L. Moore, Sharien Fitriasari, Melissa Childers, Mihaela E. Sardiu, Selene Swanson, Dai Tsuchiya, Jay Unruh, George Bugarinovic, Lin Li, Rita Shiang, Annita Achilleos, Jill Dixon, Michael J. Dixon, Paul A. Trainor
Directed differentiation of EA/TEF patient-derived induced pluripotent stem cells into esophageal epithelial organoids reveal SOX2 dysregulation at the anterior foregut stage
Suleen Raad, Anu David, Melanie Sagniez, Zakaria Orfi, Nicolas A. Dumont, Martin Smith, Christophe Faure
The impact of hyperglycemia upon BeWo trophoblast cell metabolic function: A multi-OMICS and functional metabolic analysis
Zachary JW Easton, Xian Luo, Liang Li, Timothy RH Regnault
DE-NOVO HEMATOPOIESIS FROM THE FETAL LUNG
Anthony K. Yeung, Carlos Villacorta-Martin, Jonathan Lindstrom-Vautrin, Anna C. Belkina, Kim Vanuytsel, Todd W. Dowrey, Alexandra B. Ysasi, Vladimir Vrbanac, Gustavo Mostoslavsky, Alejandro B. Balazs, George J. Murphy
Examining the effect of chronic intranasal oxytocin administration on the neuroanatomy and behavior of three autism-related mouse models
Zsuzsa Lindenmaier, Jacob Ellegood, Monique Stuive, Kaitlyn Easson, Yohan Yee, Darren Fernandes, Jane Foster, Evdokia Anagnostou, Jason P. Lerch
Acoel single-cell atlas reveals expression dynamics and heterogeneity of a pluripotent stem cell population
Ryan E. Hulett, Julian O. Kimura, D. Marcela Bolaños, Yi-Jyun Luo, Lorenzo Ricci, Mansi Srivastava
Spontaneous and ART-induced large offspring syndrome: similarities and differences in DNA methylome
Yahan Li, Jordana Sena Lopes, Pilar Coy Fuster, Rocío Melissa Rivera
Decoding the IGF1 Signaling Gene Regulatory Network Behind Alveologenesis from A Mouse Model of Bronchopulmonary Dysplasia
F Gao, C Li, SM Smith, N Peinado, G Kohbodi, E Tran, E Loh, W Li, Z Borok, P Minoo
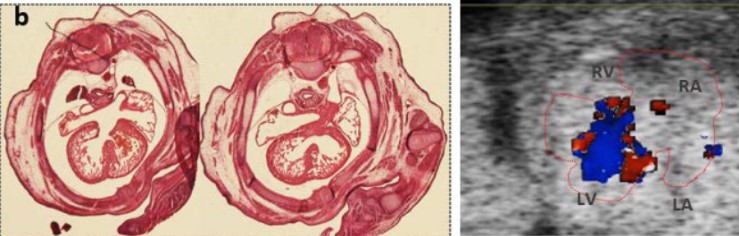
A Mouse Model with Complete Penetrance for Atrioventricular Septal Defect/Complete AV Canal
Yicong Li, Peter Andersen, Xihe Liu, Anna J. Moyer, Chulan Kwon, Roger H. Reeves
Control of Craniofacial Development by the Collagen Receptor, Discoidin Domain Receptor 2
Fatma F. Mohamed, Chunxi Ge, Randy T. Cowling, Noriaki Ono, Abdul-Aziz Binrayes, Barry Greenberg, Vesa M. Kaartinen, Renny T. Franceschi
Ablation of SAMD1 in Mice Causes Failure of Angiogenesis, Embryonic Lethality
Bruce Campbell, Sandra Engle, Terence Ozolins, Patricia Bourassa, Robert Aiello
Maternal thyroid hormone increases neural cell diversity during zebrafish spinal cord neurodevelopment
Nádia Silva, Marco António Campinho
Zebrafish her3 knockout impacts developmental and cancer-related gene signatures
Matthew R. Kent, Delia Calderon, Katherine M. Silvius, Collette A. LaVigne, Matthew V. Cannon, Genevieve C. Kendall
A novel missense mutation in the proprotein convertase gene furinb causes hepatic cystogenesis during liver development in zebrafish
Jillian L. Ellis, Kimberley J. Evason, Changwen Zhang, Makenzie N. Fourman, Jiandong Liu, Nikolay Ninov, Marion Delous, Benoit Vanhollebeke, Ian Fiddes, Jessica P. Otis, Yariv Houvras, Steven A. Farber, Xiaolei Xu, Xueying Lin, Didier Y.R. Stainier, Chunyue Yin
Nr2f1a maintains atrial nkx2.5 expression to repress pacemaker identity within venous atrial cardiomyocytes
Kendall E. Martin, Padmapriyadarshini Ravisankar, Manu E. M. Beerens, Calum A. MacRae, Joshua S. Waxman
Epidermal basal domains organization highlights skin robustness to environmental exposure
Sangeeta Ghuwalewala, Seon A Lee, Kevin Jiang, Joydeep Baidya, Gopal Chovatiya, Pritinder Kaur, David Shalloway, Tudorita Tumbar
Impaired Lef1 activation accelerates iPSC-derived keratinocytes differentiation in Hutchinson-Gilford Progeria Syndrome
Xiaojing Mao, Zheng-Mei Xiong, Huijing Xue, Markus A. Brown, Yantenew G. Gete, Reynold Yu, Linlin Sun, Kan Cao
GPX4-associated Sedaghatian Type Spondylometaphyseal Dysplasia: A Protein Interactome Perspective
Kalyani B. Karunakaran, N. Balakrishnan, Madhavi K. Ganapathiraju
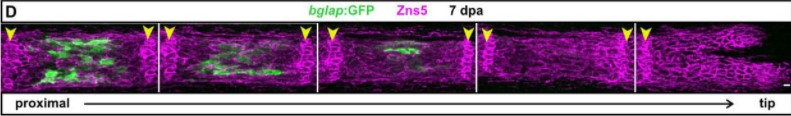
Zebrafish fin regeneration requires generic and regeneration-specific responses of osteoblasts to trauma
Ivonne Sehring, Melanie Haffner-Luntzer, Anita Ignatius, Markus Huber-Lang, Gilbert Weidinger
Quantitative Proteomic Profiling of Murine Embryonic Heart Development Reveals a Role for the Mevalonate Pathway in Cardiomyocyte Proliferation
Whitney Edwards, Todd M. Greco, Gregory E. Miner, Natalie K. Barker, Laura Herring, Sarah Cohen, Ileana M. Cristea, Frank L. Conlon
Aberrant cortical development is driven by impaired cell cycle and translational control in a DDX3X syndrome model
Mariah L. Hoye, Lorenzo Calviello, Abigail J. Poff, Nna-Emeka Ejimogu, Carly R. Newman, Jianhong Ou, Stephen N. Floor, Debra L. Silver
Differential roles of neural crest- and endothelial-derived FOXC2 in trabecular meshwork and Schlemm’s Canal in glaucomatous pathology
Pieter R. Norden, Lisa Beckmann, Raymond Fang, Naoto Ujiie, Zhen Cai, Xian Zhang, Junghun Kweon, Ting Liu, Kazushi Aoto, Susan E. Quaggin, Hao F. Zhang, Tsutomu Kume
A unique mineralizing pool of Gli1+ stem cells builds the tendon enthesis and demonstrates therapeutic potential
Fei Fang, Yang Xiao, Elazar Zelzer, Kam W. Leong, Stavros Thomopoulos
Clonal behaviour of myogenic precursor cells throughout the vertebrate lifespan
Simon M. Hughes, Roberta C. Escaleira, Kees Wanders, Jana Koth, David G. Wilkinson, Qiling Xu
Long-Term Culture of Patient-Derived Cardiac Organoids Recapitulated Duchenne Muscular Dystrophy Cardiomyopathy and Disease Progression
Vittoria Marini, Fabiola Marino, Flaminia Aliberti, Nefele Giarratana, Enrico Pozzo, Robin Duelen, Álvaro Cortés Calabuig, Rita Larovere, Tim Vervliet, Daniele Torella, Geert Bultynck, Maurilio Sampaolesi, Yoke Chin Chai
Development of a physiological insulin resistance model in human stem cell-derived adipocytes
Max Friesen, Andrew S. Khalil, M. Inmaculada Barrasa, Jacob F. Jeppesen, David J. Mooney, Rudolf Jaenisch
CRISPR-mediated correction of skeletal muscle Ca2+ handling in a novel DMD patient-derived pluripotent stem cell model
Cristina Morera, Jihee Kim, Amaia Paredes-Redondo, Muriel Nobles, Denis Rybin, Robert Moccia, Anna Kowala, Jinhong Meng, Seth Garren, Pentao Liu, Jennifer E Morgan, Francesco Muntoni, Nicolas Christoforou, Jane Owens, Andrew Tinker, Yung-Yao Lin
A regulatory network of Sox and Six transcription factors initiate a cell fate transformation during hearing regeneration in adult zebrafish
Erin Jimenez, Claire C. Slevin, Wei Song, Zelin Chen, Stephen C. Frederickson, Derek Gildea, Weiwei Wu, Abdel G. Elkahloun, Ivan Ovcharenko, Shawn M. Burgess
| Plant development
Arabidopsis histone H3 lysine 9 methyltransferases KYP/SUVH5/6 are involved in leaf development by interacting with AS1-AS2 to repress KNAT1 and KNAT2
Fu-Yu Hung, Yun-Ru Feng, Yuan-Hsin Shih, You-Cheng Lai, Keqiang Wu
The Eucalyptus grandis chloroplast proteome: leaf development and seasonal variations
Amanda Cristina Baldassi, Tiago Santana Balbuena
Spatiotemporal growth pattern during plant nutation implies fast dynamics for cell wall mechanics and chemistry: a multiscale study in Averrhoa carambola
Mathieu Rivière, Alexis Peaucelle, Julien Derr, Stéphane Douady

Progressive maturation of the root apical meristem in Arabidopsis thaliana lateral roots
Béatrice Berthet, Lotte Bald, Marion Louveaux, Alexis Maizel
Seeing light from a different angle: the effects of diffuse light on the function, structure, and growth of tomato plants
Kendra B. L. Ellertson, Gregory R. Goldsmith, Z. Carter Berry
De novo stem cell establishment in meristems requires repression of organ boundary cell fate
Antoine Nicolas, Aude Maugarny-Calès, Bernard Adroher, Liudmila Chelysheva, Yu Li, Jasmine Burguet, Anne-Maarit Bågman, Margot E. Smit, Siobhan M. Brady, Yunhai Li, Patrick Laufs
ABA signaling prevents phosphodegradation of the Arabidopsis SR45 splicing factor to negatively autoregulate inhibition of early seedling development
Rui Albuquerque-Martins, Dóra Szakonyi, James Rowe, Alexander M. Jones, Paula Duque
Spatial control of cell division by GA-OsGRF7/8 module in a leaf explains the leaf length variation between cultivated and wild rice
Vikram Jathar, Kumud Saini, Ashish Chauhan, Ruchi Rani, Yasunori Ichihashi, Aashish Ranjan
CRISPR/Cas9 gene editing uncovers the role of CTR1 and ROS1 in melon fruit ripening and epigenetic regulation
Andrea Giordano, Miguel Santo Domingo, Leandro Quadrana, Marta Pujol, Ana Montserrat Martín-Hernández, Jordi Garcia-Mas
| Evo-devo
Comparing dormancy in two distantly related tunicates reveals morphological, molecular, and ecological convergences and repeated co-option
Laurel S. Hiebert, Marta Scelzo, Alexandre Alié, Anthony De Tomaso, Federico Brown, Stefano Tiozzo
Rapamycin treatment during development extends lifespan and healthspan
Anastasia V. Shindyapina, Yongmin Cho, Alaattin Kaya, Alexander Tyshkovskiy, José P. Castro, Juozas Gordevicius, Jesse R. Poganik, Steve Horvath, Leonid Peshkin, Vadim N. Gladyshev
Fish as model systems to study epigenetic drivers in human self-domestication and neurodevelopmental cognitive disorders
Dafni Anastasiadi, Francesc Piferrer, Maren Wellenreuther, Antonio Benítez Burraco
Evolutionary changes in the chromatin landscape reshape a developmental gene regulatory network during rapid life history divergence in sea urchins
Phillip L. Davidson, Maria Byrne, Gregory A. Wray
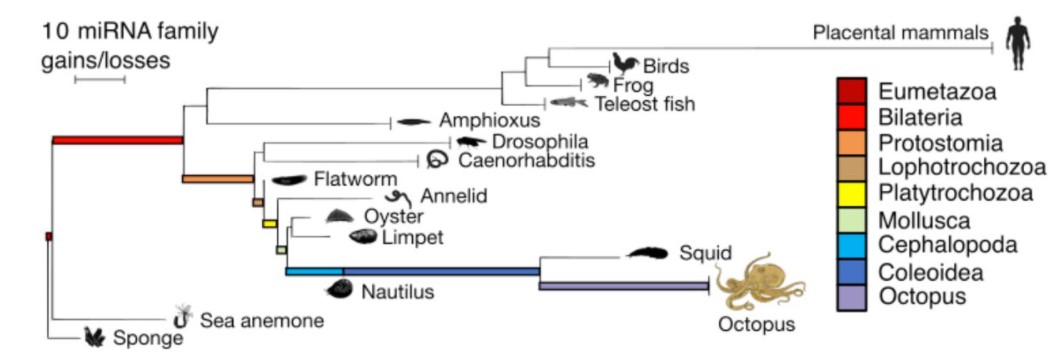
MicroRNAs are deeply linked to the emergence of the complex octopus brain
Grygoriy Zolotarov, Bastian Fromm, Ivano Legnini, Salah Ayoub, Gianluca Polese, Valeria Maselli, Peter J. Chabot, Jakob Vinther, Ruth Styfhals, Eve Seuntjens, Anna Di Cosmo, Kevin J. Peterson, Nikolaus Rajewsky
Genome editing in the unicellular holozoan Capsaspora owczarzaki suggests a premetazoan function for the Hippo pathway in multicellular morphogenesis
Jonathan E Phillips, Maribel Santos, Mohammed Kanchwala, Chao Xing, Duojia Pan
Cell Biology
Follow that cell: leukocyte migration in L-plastin mutant zebrafish.
John Bartholemew Linehan, Jose Lucas Zepeda, Taylor Ann Mitchell, Elizabeth LeClair
Kinesin-1 promotes centrosome clustering and nuclear migration in the Drosophila oocyte
Maelys Loh, Deborah Dauvet, Frida Sanchez-Garrido, Kahina Sadaouli, Fred Bernard, Antoine Guichet
Vascular mimicry by VE-cadherin enables trophoblast endovascular invasion and spiral artery remodeling during placental development
Derek C. Sung, Xiaowen Chen, Mei Chen, Jisheng Yang, Susan Schultz, Apoorva Babu, Yitian Xu, Siqi Gao, TC Stevenson Keller IV, Patricia Mericko, Michelle Lee, Ying Yang, Joshua P. Scallan, Mark L. Kahn
Maturation of cortical endoplasmic reticulum clusters in the mouse oocyte: changes at fertilization
Huizhen Wang, Lane K. Christenson, William H. Kinsey
E-cadherin mediated AMIS localisation
Xuan Liang, Antonia Weberling, Chun Yuan Hii, Magdalena Zernicka-Goetz, Clare Buckley
Cell cycle progression requires repression by Groucho during S-phase and its relief at G2-phase
Shaked Bar-Cohen, Ze’ev Paroush
Kinesin-1 promotes centrosome clustering and nuclear migration in the Drosophila oocyte
Maëlys Loh, Déborah Dauvet, Frida Sanchez-Garrido, Kahina Sadaouli, Fred Bernard, Antoine Guichet
HREM, RNAseq and cell-cycle analyses reveal the role of the G2/M-regulatory protein, Wee1, on the survivability of chicken embryos during diapause
Narayan Pokhrel, Olga Genin, Dalit Sela-Donenfeld, Yuval Cinnamon
Condensin II is required for efficient Spindle Assembly Checkpoint activation in Drosophila male meiosis
Cintia Horta, Alexandra Tavares, Raquel A. Oliveira
Crest maturation at the cardiomyocyte surface contributes to a new late postnatal development stage that controls the diastolic function of the adult heart
Clément Karsenty, Céline Guilbeau-Frugier, Gaël Genet, Marie-Hélène Seguelas, Philippe Alzieu, Olivier Cazorla, Alexandra Montagner, Yuna Blum, Caroline Dubroca, Julie Maupoint, Blandine Tramunt, Marie Cauquil, Thierry Sulpice, Sylvain Richard, Silvia Arcucci, Remy Flores-Flores, Nicolas Pataluch, Romain Montoriol, Pierre Sicard, Antoine Deney, Thierry Couffinhal, Jean-Michel Sénard, Céline Galés
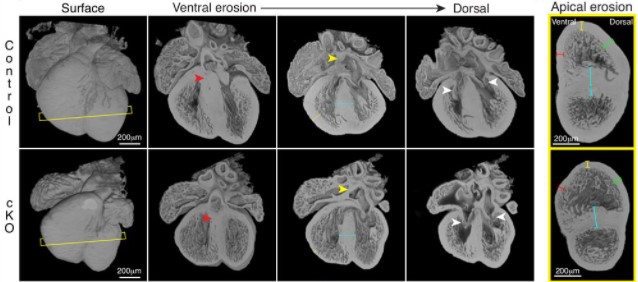
Lem2 is essential for cardiac development by maintaining nuclear integrity
Jacob A. Ross, Nathaly Arcos-Villacis, Edmund Battey, Cornelis Boogerd, Emilie Marhuenda, Didier Hodzic, Fabrice Prin, Tim Mohun, Norman Catibog, Olga Tapia, Larry Gerace, Thomas Iskratsch, Ajay M. Shah, Matthew J. Stroud
AGO1 regulates pericentromeric regions in mouse embryonic stem cells
Madlen Müller, Tara Fäh, Moritz Schaefer, Victoria Hermes, Janina Luitz, Patrick Stalder, Rajika Arora, Richard Patryk Ngondo, Constance Ciaudo
A centromeric RNA-associated protein complex affects germ line development in Drosophila melanogaster
Saskia L. Höcker, Izlem Su Akan, Alexander M. Simon, Kerem Yildirim, Lili A. Kenéz, Ingrid Lohmann, Sylvia Erhardt
Dynamically regulated Focal adhesions coordinate endothelial cell remodelling in developing vasculature
Tevin CY. Chau, Teodor E. Yordanov, Jason A. da Silva, Scott Paterson, Alpha S. Yap, Benjamin M. Hogan, Anne Karine Lagendijk
Modelling
Neutral competition within a long-lived population of symmetrically dividing cells shapes the clonal composition of cerebral organoids
Florian G Pflug, Simon Haendeler, Christopher Esk, Dominik Lindenhofer, Jürgen A Knoblich, Arndt von Haeseler
“Neighbourhood watch” model: embryonic epiblast cells assess positional information in relation to their neighbours
Hyung Chul Lee, Cato Hastings, Nidia M.M. de Oliveira, Rubén Pérez-Carrasco, Karen M. Page, Lewis Wolpert, Claudio D. Stern
Role of Delta-Notch signalling molecules on cell-cell adhesion in determining heterogeneous chemical and cell morphological patterning
Supriya Bajpai, Raghunath Chelakkot, Prabhakar Ranganathan, Mandar M. Inamdar
Automated verification, assembly, and extension of GBM stem cell network model with knowledge from literature and data
Emilee Holtzapple, Brent Cochran, Natasa Miskov-Zivanov
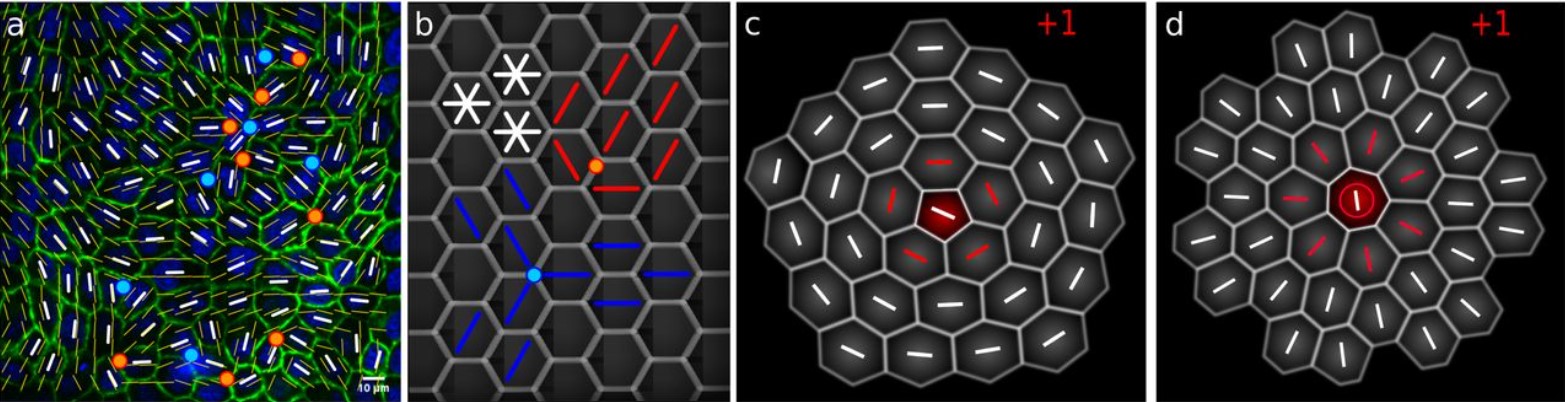
Epithelia are multiscale active liquid crystals
Josep-Maria Armengol-Collado, Livio Nicola Carenza, Julia Eckert, Dimitrios Krommydas, Luca Giomi
Flags, Landscapes and Signaling: Contact-mediated inter-cellular interactions enable plasticity in fate determination driven by positional information
Chandrashekar Kuyyamudi, Shakti N. Menon, Sitabhra Sinha
Mechanical feedback controls the emergence of dynamical memory in growing tissue monolayers
Sumit Sinha, Xin Li, Rajsekhar Das, D. Thirumalai
Stochastic fluctuations promote ordered pattern formation of cells in the Notch-Delta signaling pathway
Madeline Galbraith, Federico Bocci, José N. Onuchic
Morphology and high frequency bio-electric fields
Johann Summhammer
Oscillations and Bifurcation Structure of Reaction-Diffusion Model for Cell Polarity Formation
Masataka Kuwamura, Hirofumi Izuhara, Shin-ichiro Ei
Reviews
Control of protein-based pattern formation via guiding cues
Tom Burkart, Manon C. Wigbers, Laeschkir Würthner, Erwin Frey
Analysis and visualization of spatial transcriptomic data
Boxiang Liu, Yanjun Li, Liang Zhang
Tools & Resources
Single-Cell Multi-Omic Roadmap of Human Fetal Pancreatic Development
Sean de la O, Zhe Liu, Han Sun, Shengyang K Yu, Daniel M Wong, Emily Chu, Sneha A Rao, Nicolas Eng, Gabriel Peixoto, Jacquelyn Bouza, Yin Shen, Sarah M Knox, Aaron D Tward, Anna L Gloyn, Julie B Sneddon
Sperm morphology differences associated with pig fertility
AA Mandawala, BM Skinner, GA Walling, KE Harvey, SC Harvey
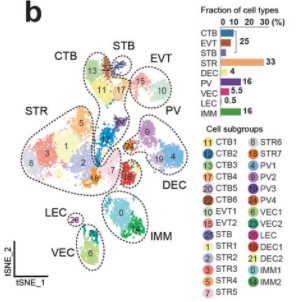
Single-cell transcriptional profiling reveals cellular and molecular divergence in human maternal-fetal interface
Quanlei Wang, Jinlu Li, Shengpeng Wang, Qiuting Deng, Yanru An, Yanan Xing, Xi Dai, Zelong Li, Qiwang Ma, Kuixing Wang, Chuanyu Liu, Yue Yuan, Guoyi Dong, Tao Zhang, Huanming Yang, Yutao Du, Yong Hou, Weilin Ke, Zhouchun Shang
Transcriptomic mapping of the metzincin landscape in human trophoblasts
Jasmin Wächter, Matthew J Shannon, Barbara Castellana, Jennet Baltayeva, Alexander G. Beristain
Single-cell characterization of neovascularization using hiPSC-derived endothelial cells in a 3D microenvironment
Simon Rosowski, Caroline Brähler, Maren Marder, Misao Akishiba, Alina Platen, Siegfried Ussar, Fabian Theis, Sandra Wiedenmann, Matthias Meier
Single-cell Bayesian deconvolution
Gabriel Torregrosa, David Oriola, Vikas Trivedi, Jordi Garcia-Ojalvo
insideOutside: an accessible algorithm for classifying interior and exterior points, with applications in embryology
Stanley E. Strawbridge, Agata Kurowski, Elena Corujo-Simon, Alexander G. Fletcher, Jennifer Nichols
TiDeTree: A Bayesian phylogenetic framework to estimate single-cell trees and population dynamic parameters from genetic lineage tracing data
Sophie Seidel, Tanja Stadler
Quantitative fate mapping: Reconstructing progenitor field dynamics via retrospective lineage barcoding
Weixiang Fang, Claire M. Bell, Abel Sapirstein, Soichiro Asami, Kathleen Leeper, Donald J. Zack, Hongkai Ji, Reza Kalhor
Cross-modality Synthesis of EM Time Series and Live Fluorescence Imaging
Anthony Santella, Irina Kolotuev, Caroline Kizilyaprak, Zhirong Bao
A single cell atlas of the cycling murine ovary
ME Morris, MC Meinsohn, M Chauvin, NMP Nguyen, HD Saatcioglu, S Yuan, A. Kashiwagi, NA. Sicher, M Hyun, PK Donahoe, B Sabatini, D Pépin
Prox1 dynamically regulates downstream targets and chromatin accessibility during venous to lymphatic endothelial cell transdifferentiation in the embryo
Lin Grimm, Elizabeth Mason, Stefanie Dudczig, Jan Kazenwadel, Tyrone Chen, Oliver Yu, Neil I. Bower, Scott Paterson, Kazuhide Okuda, Maria Rondon Galeano, Sakurako Kobayashi, Anne Senabouth, Anne K. Lagendijk, Joseph Powell, Kelly A. Smith, Natasha L. Harvey, Katarzyna Koltowska, Benjamin M. Hogan
Single cell transcriptomic and spatial landscapes of the developing human pancreas
Oladapo E. Olaniru, Ulrich Kadolsky, Shichina Kannambath, Heli Vaikkinen, Kathy Fung, Pawan Dhami, Shanta J. Persaud
High-Resolution Ultrasound and Speckle Tracking: a non-invasive approach to assess in vivo gastrointestinal motility during development
Pierre Sicard, Amandine Falco, Sandrine Faure, Jérome Thireau, Stéphanie E. Lindsey, Norbert Chauvet, Pascal de Santa Barbara
Breasi-CRISPR: an efficient genome editing method to interrogate protein localization and protein-protein interactions in the embryonic mouse cortex
Brandon L. Meyerink, KC Pratiksha, Neeraj K. Tiwari, Claire M. Kittock, Abigail Klein, Claire Evans, Louis-Jan Pilaz
A cell fate decision map reveals abundant direct neurogenesis in the human developing neocortex
Laure Coquand, Anne-Sophie Macé, Sarah Farcy, Clarisse Brunet Avalos, Amandine Di Cicco, Marusa Lampic, Betina Bessières, Tania Attie-Bitach, Vincent Fraisier, Fabien Guimiot, Alexandre Baffet
Oo-site: A dashboard to visualize gene expression during Drosophila oogenesis reveals meiotic entry is regulated post-transcriptionally
Elliot T. Martin, Kahini Sarkar, Alicia McCarthy, Prashanth Rangan
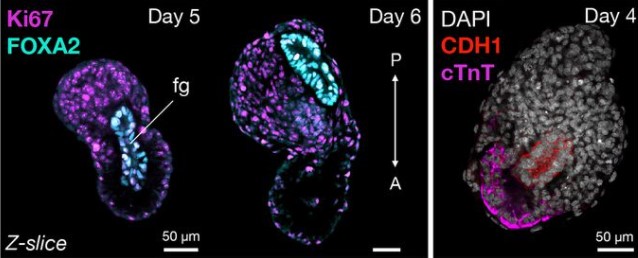
A Combined Human Gastruloid Model of Cardiogenesis and Neurogenesis
Zachary T. Olmsted, Janet L. Paluh
Stable iPSC-derived NKX2-1+ Lung Bud Tip Progenitor Organoids Give Rise to Airway and Alveolar Cell Types
Renee F.C. Hein, Ansley S. Conchola, Alexis Fine, Zhiwei Xiao, Tristan Frum, Charlie J. Childs, Yu-Hwai Tsai, Emily M. Holloway, Sha Huang, John Mahoney, Jason R. Spence
Optimization of Whole Mount RNA multiplexed in situ Hybridization Chain Reaction with Immunohistochemistry, Clearing and Imaging to visualize octopus neurogenesis
Ali M Elagoz, Ruth Styfhals, Sofia Maccuro, Luca Masin, Lieve Moons, Eve Seuntjens
Molecular Signatures and Cellular Diversity During Mouse Habenula Development
Lieke L. van de Haar, Danai Riga, Juliska E. Boer, Youri Adolfs, Thomas E. Sieburgh, Roland E. van Dijk, Kyoko Watanabe, Nicky C.H. van Kronenburg, Mark H. Broekhoven, Danielle Posthuma, Frank J. Meye, Onur Basak, R. Jeroen Pasterkamp
The Zpr-3 antibody recognizes the 320-354 region of Rho and labels both rods and green cones in zebrafish
Pan Gao, Yayun Qin, Zhen Qu, Yuwen Huang, Xiliang Liu, Jingzhen Li, Fei Liu, Mugen Liu
Bespoke data augmentation and network construction enable image classification on small microscopy datasets
Ian Groves, Jacob Holmshaw, David Furley, Benjamin D. Evans, Marysia Placzek, Alexander G. Fletcher
Single-Cell Multi-Omic Roadmap of Human Fetal Pancreatic Development
de la O Sean, Zhe Liu, Han Sun, Shengyang K. Yu, Daniel M. Wong, Emily Chu, Sneha A. Rao, Nicolas Eng, Gabriel Peixoto, Jacquelyn Bouza, Yin Shen, Sarah M. Knox, Aaron D. Tward, Anna L. Gloyn, Julie B. Sneddon
Machine learning meets classical computer vision for accurate cell identification
Elham Karimi, Morteza Rezanejad, Benoit Fiset, Lucas Perus, Sheri A. C. McDowell, Azadeh Arabzadeh, Gaspard Beugnot, Peter Siegel, Marie-Christine Guiot, Daniela F. Quail, Kaleem Siddiqi, Logan A. Walsh
Multiomic profiling defines cell fate plasticity of in vitro-derived islets
Punn Augsornworawat, Erica Marquez, Marlie M. Maestas, Matthew Ishahak, Sarah E. Gale, Mason D. Schmidt, Daniel A. Veronese-Paniagua, Julia R. Miller, Leonardo Velazco-Cruz, Jeffrey R. Millman
Dictionary learning for integrative, multimodal, and scalable single-cell analysis
Yuhan Hao, Tim Stuart, Madeline Kowalski, Saket Choudhary, Paul Hoffman, Austin Hartman, Avi Srivastava, Gesmira Molla, Shaista Madad, Carlos Fernandez-Granda, Rahul Satija
Low-cost and open-source super-resolution fluorescence microscope with autofocus for teaching and research
Justin D. Hanselman, Benjamin G. Kopek
Next Generation Opto-Jasplakinolides Enable Local Remodeling of Actin Networks
Florian Küllmer, Nynke A. Vepřek, Malgorzata Borowiak, Veselin Nasufović, Sebastian Barutzki, Oliver Thorn-Seshold, Hans-Dieter Arndt, Dirk Trauner
SpheroidAnalyseR – an online platform for analysing data from 3D spheroids or organoids grown in 96-well plates
Rhiannon Barrow, Joseph N Wilkinson, Yichen He, Martin Callaghan, Anke Brüning-Richardson, Mark Dunning, Lucy F Stead
Improved fluorescent proteins for dual-color post-embedding CLEM
Dingming Peng, Na Li, Wenting He, Kim Ryun Drasbek, Tao Xu, Mingshu Zhang, Pingyong Xu
Inference of long-range cell-cell mechanical communication from ECM remodeling fluctuations
Assaf Nahum, Yoni Koren, Bar Ergaz, Sari Natan, Shahar Goren, Avraham Kolel, Sankar Jagadeeshan, Moshe Elkabets, Ayelet Lesman, Assaf Zaritsky
DetecDiv, a deep-learning platform for automated cell division tracking and replicative lifespan analysis
Théo Aspert, Didier Hentsch, Gilles Charvin
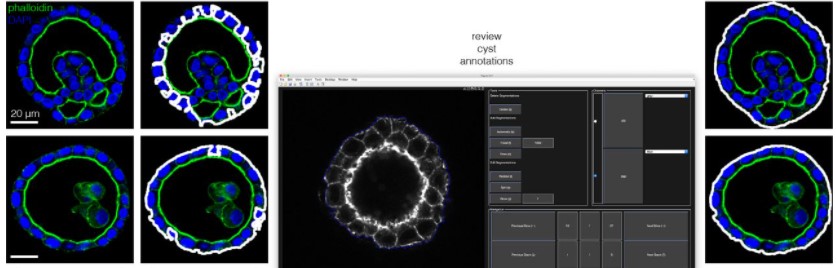
Systematically quantifying morphological features reveals constraints on organoid phenotypes
Lauren E. Beck, Jasmine Lee, Christopher Coté, Margaret C. Dunagin, Ilya Lukonin, Nikkita Salla, Marcello K. Chang, Alex J. Hughes, Joseph D. Mornin, Zev J. Gartner, Prisca Liberali, Arjun Raj
Adaptive scans allow targeted cell-ablations on curved cell sheets
Huicheng Meng, Dmitry Nuzhdin, Miguel Sison, Frédéric Galland, Loïc LeGoff
Can DyeCycling break the photobleaching limit in single-molecule FRET?
Benjamin Vermeer, Sonja Schmid
Comparative analysis of cell-cell communication at single-cell resolution
Aaron J. Wilk, Alex K. Shalek, Susan Holmes, Catherine A. Blish
MITI Minimum Information guidelines for highly multiplexed tissue images
Denis Schapiro, Clarence Yapp, Artem Sokolov, Sheila M. Reynolds, Yu-An Chen, Damir Sudar, Yubin Xie, Jeremy L. Muhlich, Raquel Arias-Camison, Sarah Arena, Adam J. Taylor, Milen Nikolov, Madison Tyler, Jia-Ren Lin, Erik A. Burlingame, Human Tumor Atlas Network, Young H. Chang, Samouil L Farhi, Vésteinn Thorsson, Nithya Venkatamohan, Julia L. Drewes, Dana Pe’er, David A. Gutman, Markus D. Herrmann, Nils Gehlenborg, Peter Bankhead, Joseph T. Roland, John M. Herndon, Michael P. Snyder, Michael Angelo, Garry Nolan, Jason R. Swedlow, Nikolaus Schultz, Daniel T. Merrick, Sarah A. Mazzilli, Ethan Cerami, Scott J. Rodig, Sandro Santagata, Peter K. Sorger
Research practice & education
Trends in Arabidopsis Research post genome sequencing- A Scientometric study
Sandeep Kumar, Amar Kant Kushwaha, R Thribhuvan, M Balakrishnan, P Krishnan
Distribution of the National Science Foundation’s Advancing Informal STEM Learning Awards (AISL) between 2006-21
Heidi M. Houzenga, Fanuel J. Muindi
Reproducibility metrics for CRISPR screens
Maximilian Billmann, Henry N. Ward, Michael Aregger, Michael Costanzo, Brenda J. Andrews, Charles Boone, Jason Moffat, Chad L. Myers
Science in motion: A qualitative analysis of journalists’ use and perception of preprints
Alice Fleerackers, Laura Moorhead, Lauren A. Maggio, Kaylee Fagan, Juan Pablo Alperin
An approachable, flexible, and practical machine learning workshop for biologists
Chris S Magnano, Fangzhou Mu, Rosemary S Russ, Milica Cvetkovic, Debora Treu, Anthony Gitter
Introducing conflict resolution and negotiation training into a biomedical sciences graduate curriculum
Michael D. Schaller, Amanda Gatesman-Ammer


 (No Ratings Yet)
(No Ratings Yet)

 (1 votes)
(1 votes)