Vitamin D: Integrating Environmental Cues into the Developmental Program
Posted by altrom, on 17 January 2019
By Amie L. T. Romney and Jason E. Podrabsky from the Podrabsky Lab at The Center for Life in Extreme Environments at Portland State University
Environmentally-induced developmental plasticity provides an opportunity to explore one of the grand challenges of modern biology – identifying mechanisms that link genotype to phenotype. The annual killifish, Austrofundulus limnaeus is a particularly useful vertebrate system to address this question. Embryos of A. limnaeus are highly resistant to environmental extremes, and may in fact define the limits for vertebrate survival without oxygen. Yet, they remain highly responsive to environmental cues during development that can trigger a remarkable capacity for plasticity. Recently, we discovered that vitamin D3 synthesis and signaling directly links environmental cues into the developmental program of this species, and supports their ability to develop along two alternative developmental trajectories. This work provides a framework for exploring the role of nuclear receptors as master regulators of developmental plasticity and life history transitions across all animal taxa.
Biology of A. limnaeus
A. limnaeus has adapted to life in a highly variable and often extreme environment. The ponds of its native habitat in Venezuela are ephemeral1, and form during the warm rainy season. High spatial and temporal variation in rain events produces ponds that are transient and can undergo multiple cycles of drying and flooding even within a single season2. When ponds dry, the entire adult and juvenile population dies, leaving only embryos to survive the dry periods3. Embryos can endure drought in a state of diapause for months while encased in the pond sediment under conditions that likely impose hypoxia/anoxia and severe desiccation stress4-6 (Figure 1). Diapause is a state of metabolic and developmental arrest that is characterized in this species by a severely depressed metabolism and heart rate, suppressed rates of proteins synthesis, the use of anaerobic pathways to support metabolism even in the presence of oxygen, and the induction of mechanisms that reduce evaporative water loss7.
There are three stages of diapause (I, II and III) possible during development in annual killifishes. Entrance into diapause II involves an alternative phenotypic trajectory that is unique morphologically, physiologically, and biochemically8, 9. Some embryos within the population bypass or “escape” diapause II and develop continuously toward hatching3, presumably completing their entire lifecycle within a single rainy season, while embryos that enter diapause II will have to wait for several months or perhaps years to hatch. Having mixed proportions of the diapause and escape phenotypes may provide an advantage for survival of the population in the face of unpredictable or fluctuating conditions. Because tolerance of extreme conditions is higher in diapausing embryos, there is a clear advantage for entering diapause II under times of extended drought.
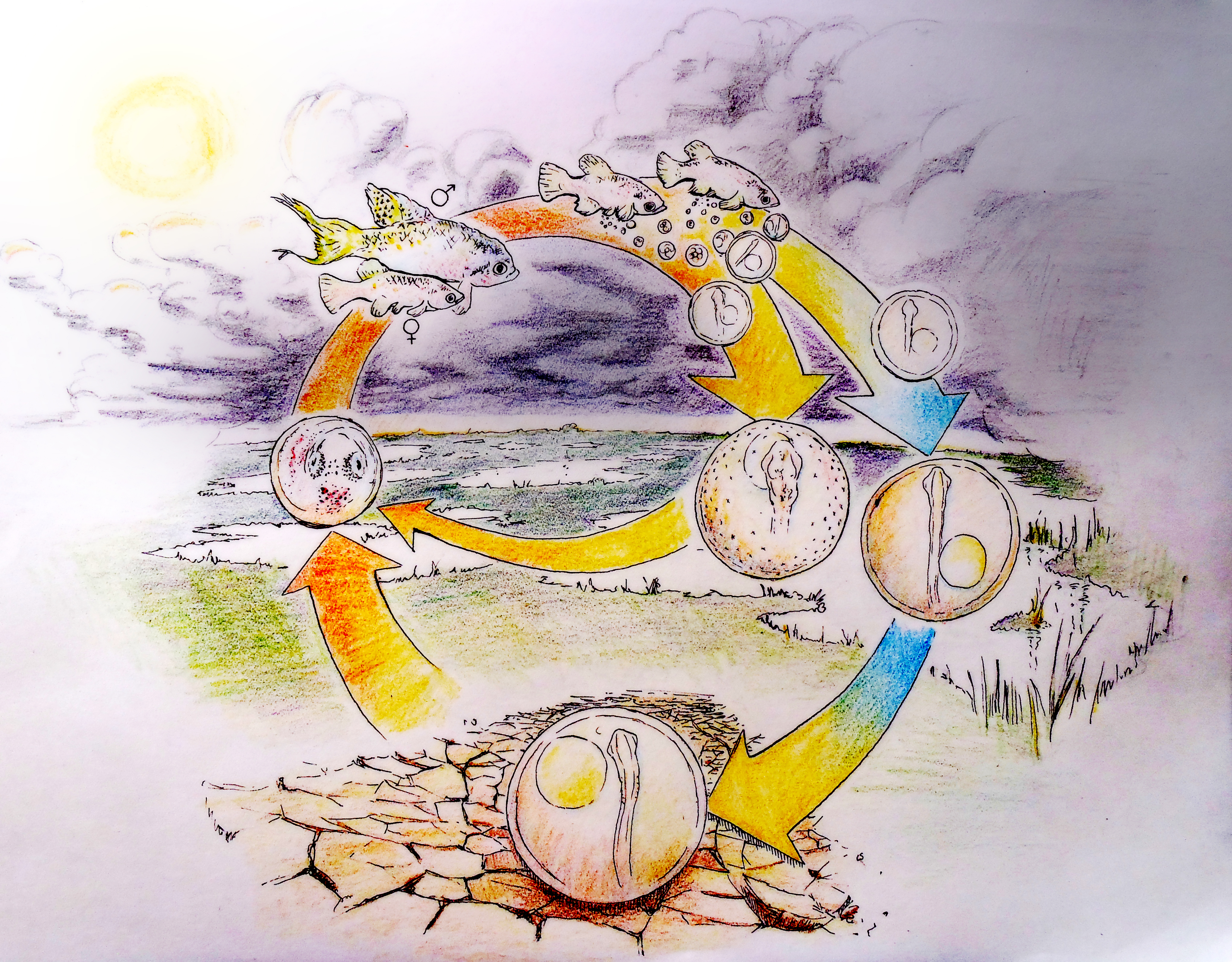
Vitamin D is not for Diapause
The developmental phenotype of killifish is regulated by two factors: an age-related maternal effect, and the incubation environment of the embryo. Typically, younger females produce a high proportion of escape embryos and older females produce diapausing embryos. However, offspring from a single spawning event, even when incubated under identical conditions, can differ in developmental outcomes. While phenotype can initially be determined by maternal patterns, the embryonic environment can override this programming9. The strongest effectors are exposure of embryos to light and incubation temperature, and there is a critical window (between 10 and 20 pairs of somites) prior to the entry into diapause II when these environmental factors cause commitment to a given developmental trajectory.
We recently identified temperature-dependent vitamin D3 signaling as a regulator of developmental phenotype in A. limnaeus10. When eggs from a single female are incubated at temperatures of 30˚C and 20˚C, embryos develop exclusively along escape and diapause trajectories, respectively, independent of maternal influences9. Under warmer conditions, embryos express a network of genes that code for enzymes responsible for synthesizing 1α, 25-dihydroxyvitamin D3, the active form of vitamin D3, and the signaling molecule that binds to the vitamin D receptor (VDR). Presumably, VDR binding to DNA response elements initiates a gene expression program that drives active growth and development along the escape trajectory (Figure 2).
Common knowledge of vitamin D3 synthesis and signaling is based largely on studies in humans and other terrestrial mammals, and spurred by the discovery of the importance of vitamin D3 in preventing rickets, a disease caused by perturbations in blood calcium homeostasis. Vitamin D3 is not actually a vitamin, but is rather a highly potent hormone that, under the right conditions, can be synthesized by humans and many other species from the precursor molecule 7-dehydrocholesterol (7-DHC). In humans, exposure of 7-DHC in the skin to UV-B light and increased temperatures results in the production of vitamin D3 (cholecalciferol)11. Vitamin D3 is then hydroxylated by two different enzymes to produce 1α, 25-dihydroxyvitamin D3. In many systems it appears that formation of vitamin D3 is a major limiting step in this synthetic pathway – thus linking seasonal changes in light and temperature to organismal levels of vitamin D3. It important to note that some species of fish can synthesize vitamin D3 using blue light and recent work suggests the potential for enzymatic conversion of 7-DHC to vitamin D3-like compounds in mammals12, 13.
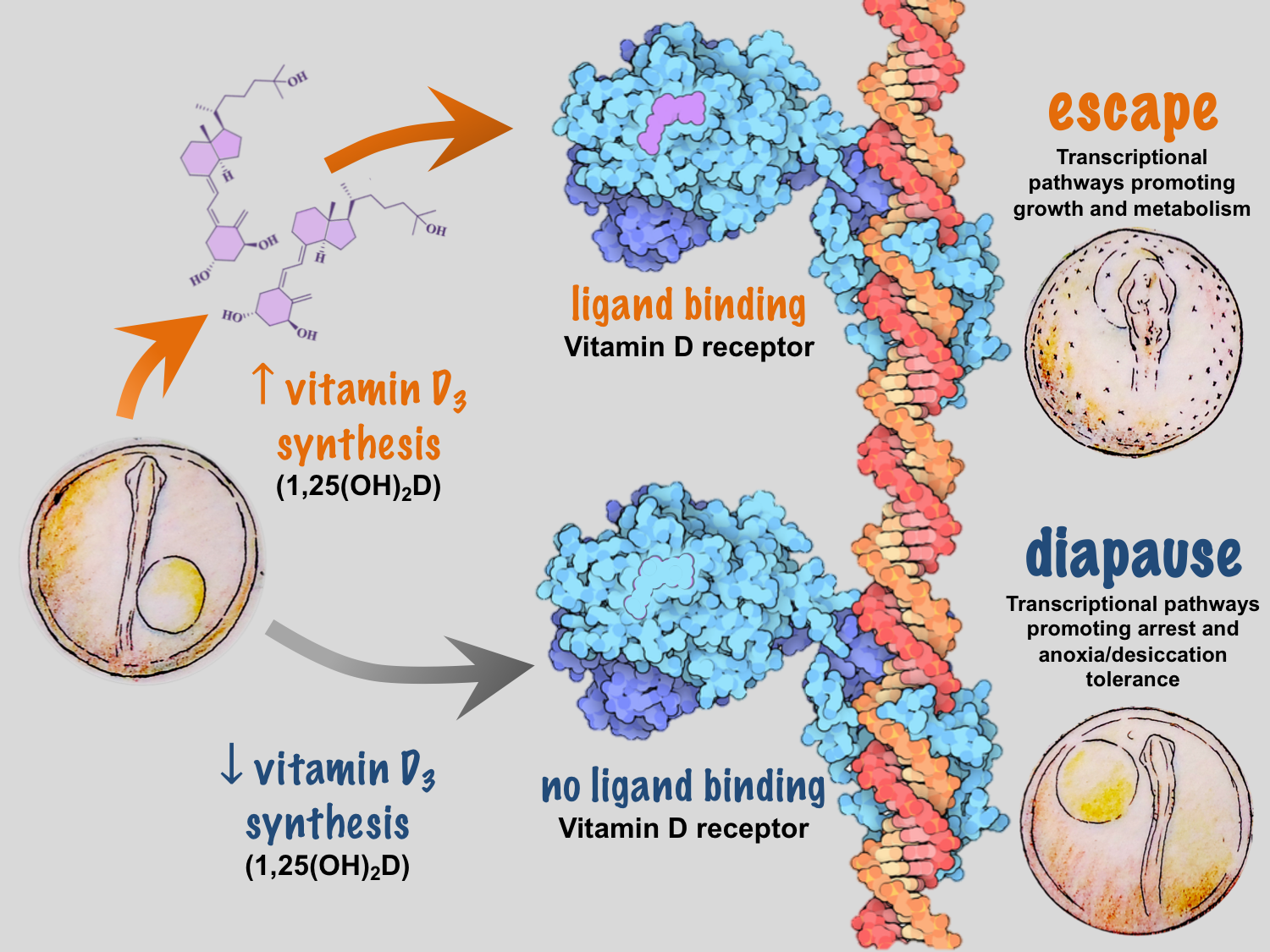
Deep conservation for regulating life history decisions in animals
The VDR is a nuclear receptor (NR) – a transcription factor whose action is regulated by binding to specific molecules or ligands. Upon ligand binding, the VDR forms heterodimers with coactivating NRs such as the retinoid X receptor and the thyroid hormone receptor to drive cascades of gene transcription for calcium transport, hormone secretion, and cellular proliferation and differentiation14. The VDR is the vertebrate homolog of daf-12, a NR critical for regulating dauer dormancy – a state of developmental arrest similar in many ways to A. limnaeus diapause – in the nematode Caenorhabditis elegans15, 16. It is well understood that environmental induction of insulin/IGF-1, TGFβ, and cGMP signaling pathways converge on DAF-12 to inhibit dauer formation and promote active growth, development, and reproduction15. Thus, it appears that regulation of developmental arrest and dormancy is regulated by homologous pathways in nematodes and fishes, suggesting a deeply conserved mechanism for integration of environmental signals into animal developmental programs.
Maternal-embryo conflict: checkpoints for phenotypic plasticity
The ecological significance of light and temperature in vitamin D3 signaling and regulation of developmental trajectory is not fully understood in A. limnaeus or any other species of annual killifish. In their native habitat in the Maracaibo basin of Venezuela, embryos of A. limnaeus are exposed to a wide range of temperatures and may – especially during the dry season – be exposed to light. Thus, vitamin D3 synthesis can provide a direct link between variations in the developmental environment and regulation of phenotype. However, developmental phenotype in A. limnaeus is regulated at two separate life stages: through maternal influences during oogenesis and later by the environmental conditions experienced by free-living developing embryos (Figure 3). Having two checkpoints that offer a regulatory capacity for determining developmental phenotype may offer a distinct survival advantage in the highly variable and often unpredictable seasonal ponds.
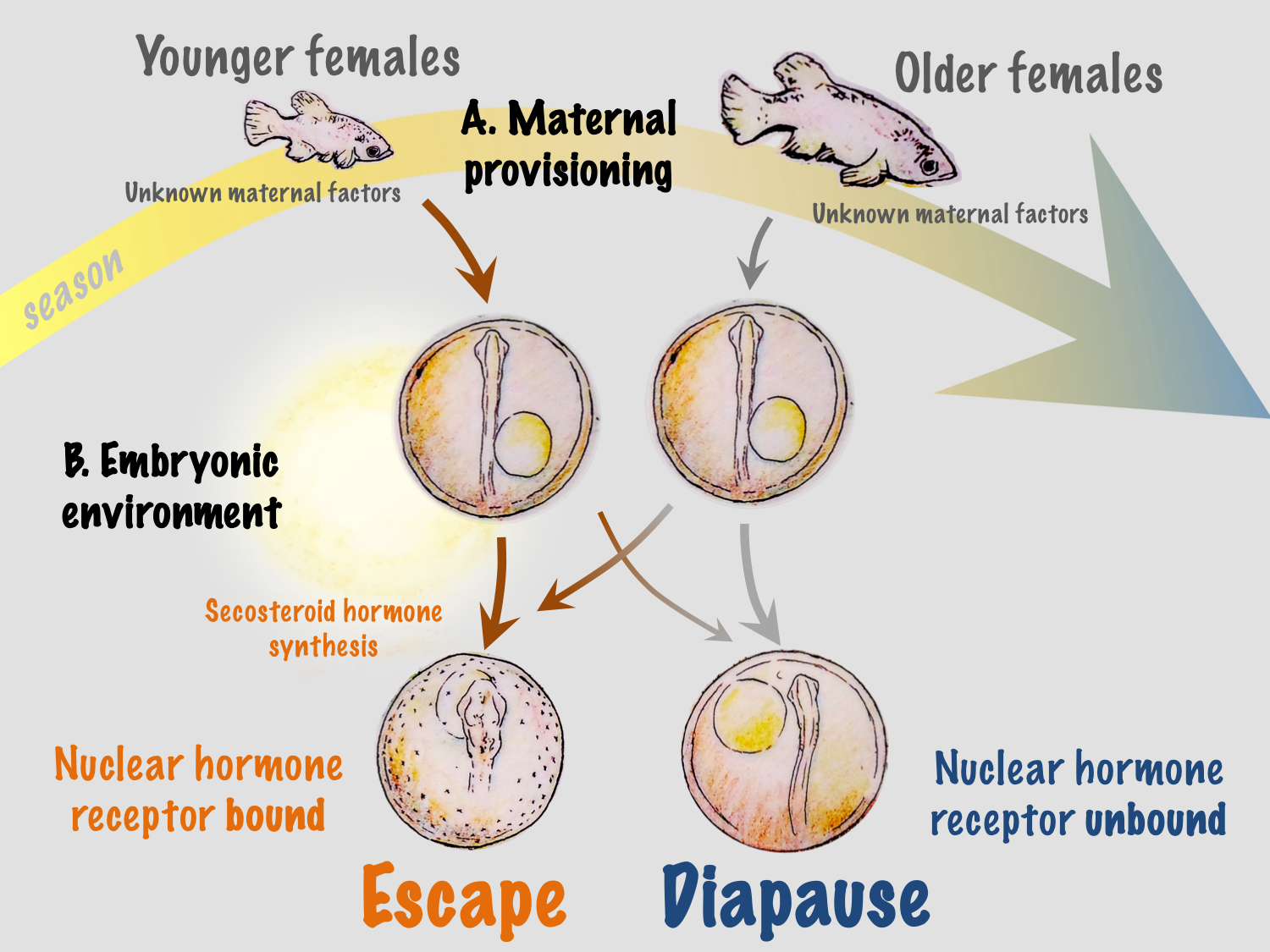
We hypothesize that maternal influences related to maternal age may help to integrate more reliable and predictable environmental conditions into the developmental program. Ecologically, this is logical because young females are usually found in newly formed ponds at the beginning of the season, and their early offspring have the greatest probability of completing the life cycle within a single rainy season. This may be especially important in ponds that experience multiple inundation and drying events during a single rainy season, allowing multiple generations or attempts at reproduction during a single season – a scenario that would be almost impossible if all embryos entered into diapause II. Maternal influences may also integrate other environmental factors that have yet to be explored in this system, such as food quality and quantity and social interactions.
Regulation of developmental phenotype during embryogenesis in a free-living embryo offers an opportunity to alter developmental outcomes in response to environmental cues that are inconsistent with maternal programming. Recent fieldwork suggests that embryos of annual killifishes remain in diapause I during the duration of the rainy season, and that development between diapause I and II occurs during the initial period of pond drying. Thus, the narrow window of development when temperature and light may affect development would allow for the embryo to “assess” environmental conditions when the ponds are drying and potentially alter developmental trajectory based on local environmental conditions. In this scenario, warm and moist conditions and/or exposure to light would result in active development while cool and dark conditions would favor entrance into diapause II. This mechanism could allow for embryos to respond to prevailing seasonal patterns in the environment, or to local microenvironmental cues. Perhaps embryos that are buried deep in the substrate remain cool and dark and enter into diapause II, while those in the upper layers experience warmer conditions with the chance for light exposure and thus develop along the escape trajectory. It is important to note that very little is known about the spawning behavior of annual killifishes, and it is possible that females may choose spawning sites, for instance in the shallow pond periphery, where the chances for exposure to higher temperatures and light are more likely. Thus, adding another layer of complexity to the potential for bet-hedging of developmental phenotype in this species.

Animals have evolved exquisite strategies to optimize developmental programs to prevailing environmental conditions to maximize growth, survival, and fitness. Here we have uncovered what appears to be a deeply conserved mechanism for integrating environmental cues into animal developmental programs with respect to entrance into developmental dormancy. This discovery suggests that many other developmental and life history transitions may be regulated in a similar manner. While it has been known for many years that nuclear receptors can be powerful regulators of phenotype, their role in developmental plasticity has received much less attention. We predict a major role for the VDR and other NRs in the regulation of other major vertebrate life history transitions such as smoltification in salmonids, metamorphosis in amphibians, and hibernation in small mammals.
1. Thomerson, J. and D. Taphorn, The annual killifishes of Venezuela part I: Maracaibo basin and coastal plain species, in Tropical Fish Hobbyist. 1992. p. 70-96.
2. Podrabsky, J.E., T. Hrbek, and S.C. Hand, Physical and chemical characteristics of ephemeral pond habitats in the Maracaibo basin and Llanos region of Venezuela. Hydrobiologia, 1998. 362:67-78. DOI: 10.1023/A:1003168704178.3. Wourms, J.P., The developmental biology of annual fishes III. Pre-embryonic and embryonic diapause of variable duration in the eggs of annual fishes. Journal of Experimental Zoology, 1972. 182:389-414. DOI: 10.1002/jez.1401820310.
4. Myers, G.S., Annual fishes. Aquarium Journal, 1952. 23:125-141.
5. Podrabsky, J.E., J.F. Carpenter, and S.C. Hand, Survival of water stress in annual fish embryos: dehydration avoidance and egg envelope amyloid fibers. American Journal of Physiology, 2001. 280:R123-R131.
6. Podrabsky, J.E., et al., Extreme anoxia tolerance in embryos of the annual killifish Austrofundulus limnaeus: Insights from a metabolomics analysis. Journal of Experimental Biology, 2007. 210:2253-2266. DOI: 10.1242/jeb.005116.
7. Podrabsky, J.E. and S.C. Hand, The bioenergetics of embryonic diapause in an annual killifish, Austrofundulus limnaeus. Journal of Experimental Biology, 1999. 202:2567-2580.
8. Podrabsky, J., A. Romney, and K. Culpepper, Alternative Developmental Pathways, in Annual Fishes. Life History Strategy, Diversity, and Evolution, N. Berois, G. García, and R. De Sá, Editors. 2016, CRC Press, Taylor & Francis: Boca Raton, FL USA. p. 63-73.
9. Podrabsky, J.E., I.D.F. Garrett, and Z.F. Kohl, Alternative developmental pathways associated with diapause regulated by temperature and maternal influences in embryos of the annual killifish Austrofundulus limnaeus. Journal of Experimental Biology, 2010. 213:3280-3288. DOI: 10.1242/jeb.045906.
10. Romney, A., et al., Temperature Dependent Vitamin D Signaling Regulates Developmental Trajectory Associated with Diapause in an Annual Killifish. Proceedings of the National Academy of Sciences of the United States of America, 2018. 115:12763-12768.
11. Holick, M.F., et al., Photosynthesis of previtamin D3 in human skin and the physiologic consequences. Science, 1980. 210:203-205.
12. Slominski, A.T., et al., Novel activities of CYP11A1 and their potential physiological significance. Journal of Steroid Biochemistry and Molecular Biology, 2015. 151:25-37.
13. Pierens, S. and D. Fraser, The origin and metabolism of vitamin D in rainbow trout. Journal of Steroid Biochemistry and Molecular Biology, 2015. 145:58-64.
14. Ramagopalan, S.V., et al., A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Research, 2010. 20:1352-1360.
15. Fielenbach, N. and A. Antebi, C. elegans dauer formation and the molecular basis of plasticity. Genes and Development, 2008. 22:2149-65. DOI: 10.1101/gad.1701508.
16. Antebi, A., et al., Daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes and Development, 2000. 14:1512-1527.


 (4 votes)
(4 votes)
 (No Ratings Yet)
(No Ratings Yet)