My name is Rob Fordham and I’ve just finished my PhD at the Wellcome Trust/MRC Cambridge Stem Cell Institute, University of Cambridge, supervised by Dr Kim Jensen (now Associate Professor at the Biotech Research and Innovation Center (BRIC), University of Copenhagen), and Professor Roger Pedersen.* During my PhD I was fascinated with intestinal development- and here I hope to convince you why this is of interest not just to developmental biologists!
The mature mammalian intestinal epithelium is comprised of villi, lined with absorptive enterocytes, protruding into the lumen, and proliferative crypt compartments invaginated into the underlying mesenchyme. The mature intestinal epithelium is the most rapidly self-renewing tissue in adult mammals, with a turnover time in the mouse of 3-4 days. We now know that this high rate of cell replenishment is driven by the division of intestinal stem cells sitting at the crypt base, intercalated between Paneth cells. This has made the intestine an excellent model system for studying tissue stem cell biology, in both homeostasis and disease. However, the emergence of this state, with functionally mature tissue stem cells, from its foetal progenitors during development was relatively poorly understood when I started my PhD.
Morphologically, we do know that by E9.0 of mouse development, a sealed gut tube has formed, which progressively lengthens over the subsequent four days. Around E14.5, the pseudostratified epithelium transitions into a simple columnar epithelium, followed by mesenchymal invagination around E15.5 to form nascent villi. Thus, in mice at birth, the intestine is developmentally immature, consisting simply of a series of undulating villi and proliferative intervillus regions. During the first two postnatal weeks, the intervillus regions lengthen into nascent crypts, coincident with the emergence of Paneth cells. Development of the human intestine follows broadly the same sequence of events as in the mouse, except that crypts start to form in utero, during the second and third trimesters. The early postnatal mouse intestine thus provides an interrogatable model system to understand the events that occur in humans during late gestation.
In 2009, methods were devised for the long-term three-dimensional culture of adult intestinal epithelium as organoids. These organoids require Wnt signalling and contain all the major cell types of the adult intestinal epithelium in vivo, including stem cells and Paneth cells (Sato et al., 2009).
By adapting these culture conditions for foetal material, we were able to demonstrate the existence of a rapidly proliferating population of intestinal progenitors that exists prior to the establishment of mature intestinal stem cells. These cells, present in the foetal intestine from E14.5, grow in vitro as cystic Foetal Enterospheres (FEnS). Through subsequent postnatal development, FEnS are replaced by organoids, and beyond around P15 only organoids can be grown. We also found that a similar population of proliferative progenitors exists in the human intestine towards the end of the first trimester (Fordham et al., 2013).
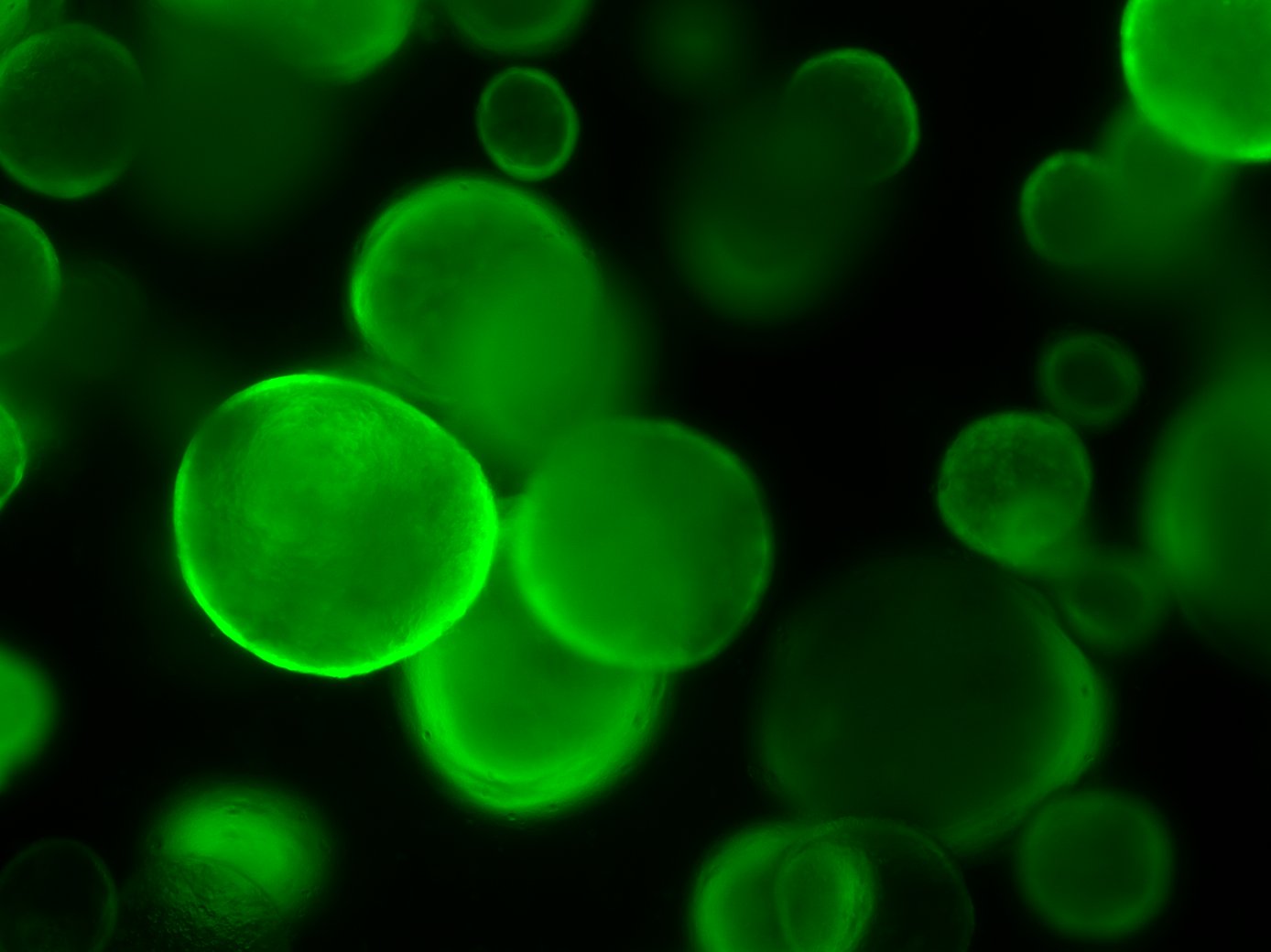 Mouse Foetal Enterospheres (mFEnS), expressing GFP, in 3D culture. From Fordham et al., Cell Stem Cell (2013)
Mouse Foetal Enterospheres (mFEnS), expressing GFP, in 3D culture. From Fordham et al., Cell Stem Cell (2013)
Thus, an otherwise developmentally transient population can be expanded in vitro, allowing direct comparison between foetal and adult intestinal stem cell populations for the first time. One important finding was that FEnS could grow without the Wnt-augmenting factor R-spondin, and even in the presence of Wnt signalling inhibitors, in stark contrast to adult organoids, which require Wnt signalling. We further demonstrated that canonical Wnt signalling is sufficient to induce the maturation of foetal intestinal progenitors into adult-like organoids (Fordham et al., 2013). Another group, from Brussels, independently described the same population of immature mouse intestinal progenitors. Mustata et al., (2013) showed that the foetal-to-adult conversion could be induced by inhibiting Notch signalling. Together these two findings suggest that the maturation of foetal progenitors proceeds in concert with establishment of the stem cell niche and that intestinal epithelial maturation requires modulation of Wnt and Notch signalling.
Of course this is all very interesting for developmental biologists, and has raised new questions for the intestine field to now address, such as the extent and relevance of heterogeneity within the pool of intervillus cells.
But I believe this also has important implications for the more translational arm of stem cell research. In particular, it is well known that most of the current protocols aiming to generate various tissue types from pluripotent cells tend to result in immature foetal-like populations, rather than functionally mature adult tissues. Indeed, we were able to generate intestinal cells from the directed differentiation of human iPS cells, but these cells displayed more foetal than adult characteristics (Hannan et al., 2013; Fordham et al., 2013). Clearly the conversion of these immature cells into an adult state in vitro will require a much better understanding of tissue stem cell specification during normal development in vivo.
Since the overarching aim of regenerative therapy is replacement of damaged tissue, we were interested in assessing the in vivo potential of immature intestinal cells. Importantly we now had a system, in the form of mouse FEnS, to sufficiently expand immature cells for transplantation experiments. Previous work from Tokyo Medical and Dental University had shown that adult colonic organoids, transplanted into mice with damaged colons, could contribute to regeneration of the colonic epithelium (Yui et al., 2012). Working with the group in Tokyo, we were able to demonstrate that mouse FEnS could similarly contribute to colonic epithelial regeneration following transplantation (Fordham et al., 2013). This proof-of-principle experiment revealed that foetal cells cultured for prolonged periods in 3D culture retain the potential for differentiation into more-mature cell types, perhaps with greater lineage plasticity than adult-derived cells.
Therefore this work should highlight why translational stem cell research should not lose sight of basic developmental biology- and that developmental biology is perhaps not so far removed from translational impact as people think!
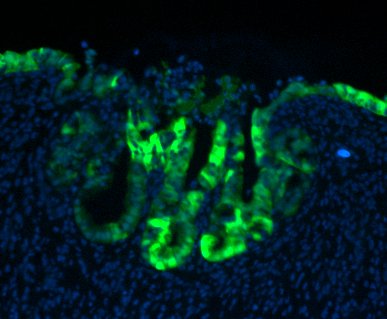 GFP-expressing mouse FEnS, transplanted into a damaged colon, contribute to epithelial regeneration
GFP-expressing mouse FEnS, transplanted into a damaged colon, contribute to epithelial regeneration
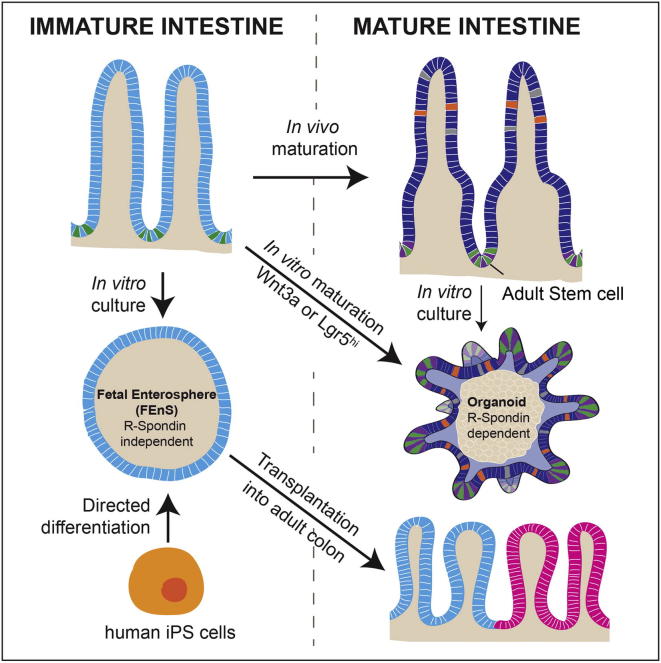 Summary of the major findings presented in Fordham et al., Cell Stem Cell (2013)
Summary of the major findings presented in Fordham et al., Cell Stem Cell (2013)
References
Fordham, R.P. and Yui, S. et al., Transplantation of Expanded Fetal Intestinal Progenitors Contributes to Colon Regeneration after Injury. Cell Stem Cell (2013) Dec 5; 13 (6) 734-744
Hannan, N.R.F., Fordham, R.P. et al., Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Reports (2013) Oct 10; 1 (4); 293-306
Miyoshi, H. and Stappenbeck, T.S. The Young and the Wnt-less: Transplantable Fetal Intestinal Spheroids without Wnts. Cell Stem Cell (2013) Dec 5; 13 (6) 637-638
Mustata, R.C. et al., Identification of Lgr5-independent spheroid-generating progenitors of the mouse fetal intestinal epithelium. Cell Rep. (2013) Oct 31;5(2):421-32
Sato, T. et al., Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature (2009) May 14;459(7244):262-5
Yui, S. et al., Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. (2012) Mar 11;18(4):618-23
This work was funded by the Medical Research Council and Wellcome Trust
*from January 2014, I will be a Postdoctoral Research Fellow in Professor Owen Sansom’s group at The Beatson Institute for Cancer Research, Glasgow
 (4 votes)
(4 votes)
 Loading...
Loading...



 (No Ratings Yet)
(No Ratings Yet)

 (2 votes)
(2 votes)







 (22 votes)
(22 votes)



