Development editorial- Developing with the community
Posted by Katherine Brown, on 17 December 2013
The following editorial, by Development’s Editor in Chief Olivier Pourquié, was first published in Development.
In recent years, there has been much discussion and some degree of discontent with the current scientific publishing system. While we at Development continue to believe in the basic tenets of academic journal publishing – stringent editorial assessment and pre-publication peer review – we recognise that we can always improve, and that we should strive to serve our community as best we can. 2013 has seen some changes to the publishing policies of Development (more on which below), and these will continue in the coming year. With new technologies altering the way scientists access and digest research, and with the changes in the developmental biology field – expanding into stem cell science, quantitative biology and other areas – we also need to engage with and expand our community to reflect these changes.
As many of you will be aware, 2013 was a big year for Development in the stem cell field. As a leading journal for developmental biologists, we believe that it is key to maintain and strengthen the links between the stem cell and developmental fields, and we have continued our efforts to reach out to the stem cell community and encourage scientists in this area to publish their best work in Development. Notably, in 2013, we created a new website, ‘Stem Cells and Regeneration’ (stemcells.dev.biologists.org), which gathers together all the stem cell papers published in the journal and provides a simple ‘one-stop shop’ for stem cell scientists; it also includes community news and useful links. The website was launched at the ISSCR Annual Meeting in Boston in June, along with a Special Issue of the journal dedicated to stem cells and regeneration. We are particularly proud of this issue, which gathers excellent opinion pieces and reviews from prominent stem cell scientists and developmental biologists, and which has been enthusiastically received by old and new readers alike. In time, we would like stem cell researchers to come to view Development as their community journal, as we hope that developmental biologists already do. Although it is still early days to evaluate the impact of these various efforts on the journal, we nevertheless have observed very encouraging trends. Notably, papers in the stem cell section of the journal top our lists of most read and most cited articles, and submissions from stem cell scientists continue to increase.
We are also trying to actively promote studies on human development in the journal. Later this year, Austin Smith, Benoit Bruneau and I are organising a Company of Biologists workshop ‘From Stem Cells to Human Development’. We have lined up a spectacular list of speakers and this should be a very exciting meeting – we hope some of you will be able to attend! For more information on this workshop, please see workshops.biologists.com/workshop_sept_2014.html.
This year has seen significant changes in the internal Development community, with two editors stepping down and being replaced, and with changes to the in-house team. Alex Joyner and Shin-Ichi Nishikawa retired from the team of academic editors and we thank them for their great work and support to the journal. We were thrilled to welcome François Guillemot from the National Institute for Medical Research (London, UK) and Benoit Bruneau from the Gladstone Institute (UCSF, USA) as their replacements. François’ work focuses on mouse forebrain development and the regulation of neural lineage. Benoit is a renowned expert in cardiac development who uses both in vivo and stem cell culture approaches to understand gene regulation in the heart. Benoit is also very active on social networks, where Development has also been expanding its profile and community. We have now a facebook page (www.facebook.com/developmentjournal) and are very active on Twitter (@Dev_journal) where you can get updates on our latest content and follow conferences attended by our Executive Editor Katherine Brown and Reviews Editors Seema Grewal and Caroline Hendry. Caroline is our recently recruited Associate Reviews Editor for the stem cell field, who trained with Melissa Little in Australia and with Ihor Lemishka in New York, and who is now very actively involved in our expansion into stem cell science.
In addition to our presence on social media sites, I hope you all know about the Node (thenode.biologists.com), our community blog for developmental biologists. Having done a fantastic job of setting up and running the site for the last three years, Eva Amsen has moved on to new challenges, and the Node is now in the capable hands of Catarina Vicente. The site goes from strength to strength, and I would in particular encourage you to look at our recent series of posts on science outreach (thenode.biologists.com/tag/outreach/) and on ‘A day in the life…’ of labs working on different model organisms (thenode.biologists.com/tag/a-day-in-the-life/). We’re always looking for contributions to the Node: all you need to do is register and get writing!
In addition to these changes to the Development editorial team, we have undertaken a complete overhaul of our editorial board to better reflect the current scope of the journal – the new board can be found on our website (dev.biologists.org/site/misc/edboard.xhtml). As well as expanding our coverage of the stem cell field, we have also recruited new members with a strong background in mathematics and physics to help promote more quantitative approaches in our field, as well as strengthening our representation in fields such as evo-devo and neurobiology. We plan to solicit advice from editorial board members more actively than in the past when considering the suitability of papers for the journal, as well as for other important strategic decisions.
As you can see from the journal content, the new emphasis on stem cell science has not detracted from the more traditional developmental biology field, where we continue to publish exciting research. We are also happy to see that we now receive a steady stream of papers in the evo-devo and quantitative biology fields, which were also recognised as priorities for the journal. The ‘Techniques and Resources’ section of the journal is also growing. Launched in 2011, the purpose of this new section is to publish the description of new techniques or resources such as databases of interest for wide communities of developmental biologists. We are delighted to see that this section has met with a great enthusiasm among the community and its articles are among the most viewed of the journal.
An important aspect of our journal is that it relies on academic editors, who are specialists in their fields, working for a not-for-profit charity: The Company of Biologists. Profits made by the journal are reinvested in the community and serve to support travel fellowships for young researchers and meeting grants to developmental biology societies and other organisations around the world (see www.biologists.com/grants.html), as well as to organise a valuable series of workshops (workshops.biologists.com/index.html). We are proud to be able to support members of our community in this way. We offer our authors a range of options for publication: you can choose either to publish your manuscript under our Open Access option and make it immediately free for the community, or you can publish under our subscription model, in which case there are no publication charges at all, and the article is free to read after 6 months. Moreover, we strive to serve our community by implementing a fair and efficient peer-review process. At present, our average time from initial submission to issue publication is 6 months, and papers rarely go through protracted rounds of revision and re-review. There is a strong commitment on our part at first decision stage, with well over 90% of papers on which a revision is invited being accepted for publication. This implies a serious evaluation of the suitability of the paper at the time of submission and first review, an effort in which our new editorial board will be more actively involved. We have also begun to share reviews on a particular manuscript among the referees in cases where this will help the editor to come to a more balanced decision.
We recognise that publishing is a highly competitive endeavour, and that authors can suffer when their work is scooped by their competitors. To help alleviate this problem, we have now implemented a policy of ‘Scoop protection’: if a competing paper comes out once we have invited a revision on a paper, we won’t reject on grounds of conceptual advance. Researchers in fields such as computational biology are increasingly turning to pre-print servers such as arXiv for the pre-publication deposition of their manuscripts – where they can be viewed by the community at an early stage in the publication process. Although this is not yet common practice in our field, there are members of our community who do make use of such servers, and we believe this number may well grow. We are therefore pleased to announce that we will not consider posting of an article on a pre-print server as prior publication, and would still consider such manuscripts for potential publication inDevelopment. Finally, on the discussion of publishing policies, we are also actively involved in the discussion on journal metrics initiated by the San Francisco Declaration on Research Assessment to limit the use of impact factor in science evaluation and promote the use of a wider panel of more objective measures. To view the text of the declaration and learn more about the initiative you can consult the editorial I wrote on the topic in May 2013 (dev.biologists.org/content/140/13/2643).
I would like to conclude by thanking the board of The Company of Biologists, particularly directors past and present, John Gurdon and Tim Hunt. I am also grateful to theDevelopment Advisory Group: James Briscoe, Cheryll Tickle and Kate Storey, for valuable discussions and support. The team of academic editors deserves great credit for their dedication and enthusiasm for the job, and I thank our editorial board for their engagement and support. I also thank the Development staff: Administrators Jenny Ostler and Debbie Thorpe; Production Editors Colin Davey, Jane Gunthorpe and Lindsay Roberts; the in-house editorial team of Katherine Brown, Seema Grewal, Caroline Hendry and Catarina Vicente; as well as the company’s production department and Publisher Claire Moulton.
Olivier Pourquié, Development Editor in Chief


 (3 votes)
(3 votes)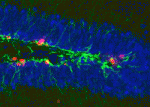 Neurotransmitter functions are typically associated with neural function rather than with development, but there is a growing body of evidence suggesting that neurotransmitters may also regulate cell proliferation and differentiation in the nervous system. Recent work has demonstrated that GABAA receptor signalling can control adult neurogenesis in the mouse. Verdon Taylor and co-workers now investigate a role for GABAB receptors in the adult mouse hippocampus (
Neurotransmitter functions are typically associated with neural function rather than with development, but there is a growing body of evidence suggesting that neurotransmitters may also regulate cell proliferation and differentiation in the nervous system. Recent work has demonstrated that GABAA receptor signalling can control adult neurogenesis in the mouse. Verdon Taylor and co-workers now investigate a role for GABAB receptors in the adult mouse hippocampus (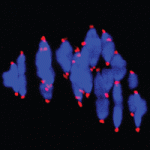 Chromosome mis-segregation during meiosis leads to aneuploidy, and hence to reduced fertility and birth defects. The frequency of aneuploidy increases in older mothers. Various mechanisms have been proposed to account for this correlation between maternal age and chromosome segregation defects, most placing the emphasis on errors occurring in meiosis I (MI). Keith Jones and colleagues (
Chromosome mis-segregation during meiosis leads to aneuploidy, and hence to reduced fertility and birth defects. The frequency of aneuploidy increases in older mothers. Various mechanisms have been proposed to account for this correlation between maternal age and chromosome segregation defects, most placing the emphasis on errors occurring in meiosis I (MI). Keith Jones and colleagues ( Vertebrate somites form via a sequential process of segmentation, which proceeds from anterior to posterior. In many species, anterior somites form more quickly than posterior ones, but whether this is functionally important, and how the switch in timing might be controlled, is unknown. Now (
Vertebrate somites form via a sequential process of segmentation, which proceeds from anterior to posterior. In many species, anterior somites form more quickly than posterior ones, but whether this is functionally important, and how the switch in timing might be controlled, is unknown. Now (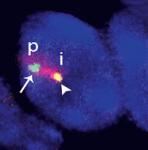 Within the nucleus, active gene loci tend to cluster together in the nuclear interior, while inactive genes are often found at the nuclear lamina. On
Within the nucleus, active gene loci tend to cluster together in the nuclear interior, while inactive genes are often found at the nuclear lamina. On 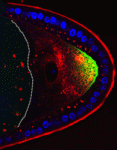 The plus-end directed motor protein Kinesin-1 is a major effector of microtubule-mediated transport, moving a wide range of cargo around cells. How cargo specificity is achieved and how motor transport is regulated are still not fully understood, particularly in in vivo developmental contexts. Isabel Palacios and colleagues (
The plus-end directed motor protein Kinesin-1 is a major effector of microtubule-mediated transport, moving a wide range of cargo around cells. How cargo specificity is achieved and how motor transport is regulated are still not fully understood, particularly in in vivo developmental contexts. Isabel Palacios and colleagues (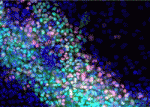 The ability to manipulate genomes has been significantly advanced by the development of the CRSIPR/Cas9 technology, in which the Cas9 endonuclease is targeted to specific sites in the genome by single-guide RNAs (sgRNA) – short sequences complementary to the genomic locus of interest. This approach has been further developed to allow regulation of gene expression at particular loci (a system referred to as CRISPRe): a catalytically inactive version of Cas9 (dCas9) can be fused to transcription activation (e.g. VP64) or repression (e.g. KRAB) domains and targeted to the desired genomic locus by sgRNA. Now, René Maehr and colleagues (
The ability to manipulate genomes has been significantly advanced by the development of the CRSIPR/Cas9 technology, in which the Cas9 endonuclease is targeted to specific sites in the genome by single-guide RNAs (sgRNA) – short sequences complementary to the genomic locus of interest. This approach has been further developed to allow regulation of gene expression at particular loci (a system referred to as CRISPRe): a catalytically inactive version of Cas9 (dCas9) can be fused to transcription activation (e.g. VP64) or repression (e.g. KRAB) domains and targeted to the desired genomic locus by sgRNA. Now, René Maehr and colleagues (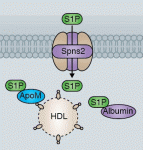 Sphingosine 1-phosphate (S1P) is a lipid mediator formed by the metabolism of sphingomyelin. Recent studies have shown that crucial events during embryogenesis, such as angiogenesis, cardiogenesis, limb development and neurogenesis, are regulated by S1P signalling. Here, Timothy Hla and colleagues provide an overview of S1P signalling in development and in disease.
Sphingosine 1-phosphate (S1P) is a lipid mediator formed by the metabolism of sphingomyelin. Recent studies have shown that crucial events during embryogenesis, such as angiogenesis, cardiogenesis, limb development and neurogenesis, are regulated by S1P signalling. Here, Timothy Hla and colleagues provide an overview of S1P signalling in development and in disease.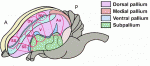 The human cerebral cortex is generally considered the most complex organ, and is the structure that we hold responsible for the repertoire of behavior that distinguishes us from our closest living and extinct relatives. At a recent Company of Biologists Workshop, ‘Evolution of the Human Neocortex: How Unique Are We?’ held in September 2013, researchers considered new information from the fields of developmental biology, genetics, genomics, molecular biology and ethology to understand unique features of the human cerebral cortex and their developmental and evolutionary origin.
The human cerebral cortex is generally considered the most complex organ, and is the structure that we hold responsible for the repertoire of behavior that distinguishes us from our closest living and extinct relatives. At a recent Company of Biologists Workshop, ‘Evolution of the Human Neocortex: How Unique Are We?’ held in September 2013, researchers considered new information from the fields of developmental biology, genetics, genomics, molecular biology and ethology to understand unique features of the human cerebral cortex and their developmental and evolutionary origin. (No Ratings Yet)
(No Ratings Yet)
 (13 votes)
(13 votes)