If I shut my eyes, I can still picture the young boy dressed as a gravedigger. He’s taking centre-stage, flourishing a spade and cheerily telling his audience that if they don’t practise safe sex, they will die of AIDS and boost his business. It’s not the sort of thing you expect from a Sunday school play.
It was February 2002 and I was in Kenya, covering a story for New Scientist magazine about how traditional African theatre could help teach local people about HIV, food hygiene and environmental threats such the pollution of nearby Lake Victoria. The science conveyed in the plays was basic, but it had the power to transform people’s lives.
That reporting assignment, which also took me to Nairobi’s slums and into the African Rift Valley to meet the Maasai people, is probably the most challenging of my career. It taught me how much you can achieve with nothing more than a notebook, pen and a pocket camera. It also sealed my conviction that sharing scientific knowledge is as valuable an enterprise as generating it.
I certainly didn’t start out with the idea of becoming any kind of journalist. The developmental biology bug bit during my undergraduate degree and I decided to pursue a career in research. Funded by a Prize Studentship from the Wellcome Trust, I joined Helen Skaer’s lab, then at the Department of Human Anatomy in Oxford, in 1995. Under Helen’s excellent tutelage, I started work on finding out how the cells of the Drosophila renal system, the Malpighian tubules, decide their fate.
But while I was happily dissecting embryos, two things changed the course of my career. The first was a science communication course run by the Wellcome Trust for its Prize students. One of the course tutors, Peter Evans, a science radio journalist for BBC Radio 4, encouraged me to try my hand at student radio. Before long, I was a writer and presenter for The Frontier, a science magazine show on Oxygen 107.9FM, the UK’s first student station with a full FM radio licence.
I have no idea how many listeners The Frontier team had, but producing a live half-hour show every week, whilst also studying for our degrees, was both hugely stressful and immensely fun. It planted the idea that this might be something I could do in future. To test the waters a bit more, I entered a number of science writing competitions, and was a runner up in the Wellcome Trust / New Scientist Essay competition.
This prompted the second career-changing event: as I was writing up my doctoral thesis, I applied for an internship as a subeditor at New Scientist. I succeeded in getting the job and joined the magazine as soon as my lab work was complete.
Working on a magazine was worlds away from counting Malpighian tubule cells, but the experience of working on The Frontier, as well as the writing competitions, did help–both in terms of doing the job and getting it in the first place.
So if I could only give one bit of advice, it would be this: if you want to get into science journalism, just do it. Take every opportunity you can–blogging, writing competitions, student newspapers or radio, writing for trade or academic publications, working as an intern–to flex your writing muscles and build a portfolio of examples to show prospective employers. Plenty of people say they want to be science journalists, but far fewer demonstrate the initiative and nous required to make it happen.
Subeditors are unsung diamond-polishers of the publishing world; their job is to edit prose for clarity, good grammar and style and then write eye-catching headlines. Being a trainee sub taught me a great deal about how to write well and fueled my desire to become a reporter / writer myself. I started writing pieces for the magazine and within a few months, I had wangled a job as a reporter in New Scientist’s news section.
I stayed in News for about 18 months before finding my métier as a feature writer, and eventually I became a features editor. Features are longer articles that give you the time and space to explore ideas in more depth and craft an article into more of a story, both of which appealed to me. In 2005, I left New Scientist to join Nature as a senior reporter and editor, where I focused on developing the biology features in Nature’s news section.
While this was all happening, I had been using my annual leave to train scientists in science communication skills, with my marine biologist husband, Jon Copley. There is an increasing demand for researchers to communicate with the public and to demonstrate the impact of their work. So Jon and I founded a company called SciConnect to offer training courses in these areas to scientists who need them.
By the time I had been at Nature for two years, the demands of SciConnect were growing. What’s more, I found that I was spending most of my time editing rather than writing. Much as a enjoyed working at Nature, I decided to leap into the unknown and become a freelance science journalist and full-time managing director of SciConnect. After I handed in my notice, I lay awake all night, fretting about whether I had just made the worst mistake of my career.
I need not have worried. Running my own company and freelancing as a writer has been terrifying, exhilarating, overwhelming, and empowering, in turn. It demands a whole new skill set and the learning curve has been precipitous. But it has its rewards: SciConnect has now equipped more than 1600 scientists, from PhD students to Profs, with the skills to share their work with the world.
There are days of doubt, of course. Every now and then I fish a sozzled fruit fly out of my Rioja, dry its bright wings and feel a pang of nostalgia for the lab. Or I’ll encounter a snotty academic dinosaur (thankfully a rare species) who thinks that “journalist” means “imbecile” and that leaving the research track somehow equates with failure or “dropping out”.
At times like these I remember the jolly little gravedigger and remind myself that science is not just about making discoveries, but also holding science to account and making sure those discoveries reach beyond the lab.
If anyone is contemplating making the leap into journalism, check out the career information on the Association of British Science Writers’ website.
Alternatively drop me a line via email (info@sciconnect.co.uk), on Twitter (@ClaireAinsworth) or my blog and I’ll do my best to help.
 (13 votes)
(13 votes)
 Loading...
Loading...
 Last week I attended the 18th international C. elegans meeting at UCLA, organised by the Genetics Society of America. Having done most of my scientific training with mammalian cell culture, I had never been to an organism-specific meeting – let alone one about worms – and I was curious to find out what it would be like. I also wanted to learn more about the sense of community that exists among worm researchers, and what better way to experience that than by visiting their conference.
Last week I attended the 18th international C. elegans meeting at UCLA, organised by the Genetics Society of America. Having done most of my scientific training with mammalian cell culture, I had never been to an organism-specific meeting – let alone one about worms – and I was curious to find out what it would be like. I also wanted to learn more about the sense of community that exists among worm researchers, and what better way to experience that than by visiting their conference.

 (4 votes)
(4 votes)

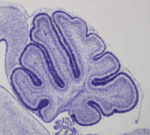
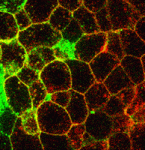
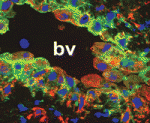
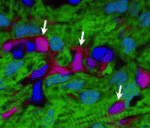
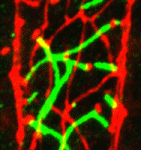
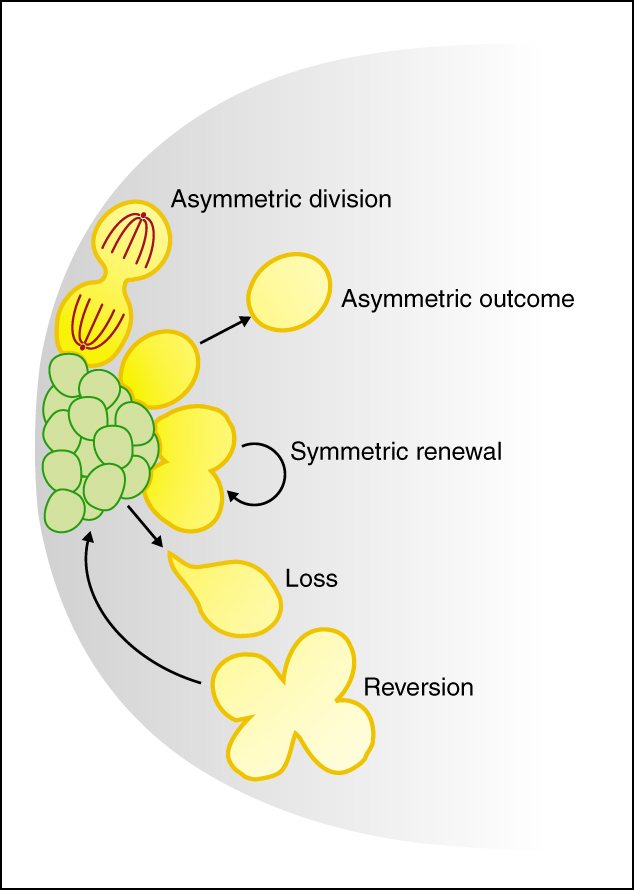 The stem cell niche: lessons from the Drosophila testis
The stem cell niche: lessons from the Drosophila testis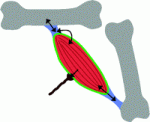
 (2 votes)
(2 votes)