The 2011 Wellcome Image Awards were announced a few weeks ago, and developmental biology is well-represented in this year’s gallery, with images featuring cell division in plants, fish eye development, blastocysts, a developing mouse kidney, chromatin density in chromosomes, caterpillar prolegs, and a mouse embryo animation.
On February 23rd, the awards were announced at an official ceremony in London. “The awards ceremony was absolutely delightful”, according to Agnieszka Jedrusik, whose blastocyst image was amongst the winners, “Dr Laura Pastorelli should be congratulated on her amazing organizational skills. There were over 200 guests invited, yet, she still made an effort to greet all guests in person and made sure they felt welcomed.” Monica Folgueira, who is behind the winning cavefish image, also enjoyed the ceremony: “Everything went very quick for me. What I liked the most was having the opportunity of meeting the organizers and other creators.” Monica’s former lab mate Kara Cerveny was also amongst the winners, but she, unfortunately, couldn’t attend: “I’m now working at Cell as a scientific editor”, Kara writes, “and couldn’t make it back to London, but I heard that the ceremony was lots of fun.”
The Node spoke to Monica, Agnieszka and Kara to find out a bit more about them and their winning images. Click any of the thumbnail images to go to the high-resolution image in the Wellcome Image database.
 Monica Folgueira
Monica Folgueira
“I’m a lecturer at University of A Coruña (Spain). I just moved back to Spain last September after a postdoc at Steve Wilson’s lab (UCL). Now that I’m back in Spain, I will try to continue studying different aspects of the development and anatomy of neuronal circuits in the zebrafish brain, in collaboration with Steve’s lab. In addition, I’m interested in brain diversity and evolution. So I plan to perform studies of comparative neuroanatomy in a few species of fish, including cavefish.
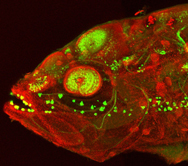
My image is a confocal micrograph of a cavefish embryo at around five days post-fertilisation. The embryo has been stained with an antibody against a calcium-binding protein (in green) to show different neuronal types and their processes in the nervous system, and with an antibody against a component of tight junctions (zona occludens- 1, in red)
I produced this image during my postdoc at Steve’s lab (UCL). This image was produced after a set of experiments whose aim was to compare the morphology of the telencephalon between various teleost fishes (including zebrafish, cavefish and medaka).
I decided to submit this image because for me it brings together some kind of beauty and drama. I find it striking that, although it is an embryo, the combination of small eyes and strong jaws makes it look more like an adult fish. The image also reveals interesting characteristics of the anatomy in fishes, like the presence of taste buds outside the oral cavity.”
 Agnieszka Jedrusik
Agnieszka Jedrusik
“I gained my first degree (BSc in 2004 followed by MSc degree in 2006) in Developmental Biology from Warsaw University, in professor Marek Maleszewski group, investigating the nature of sperm activating factor during fertilization of the mouse oocyte. In 2006 I moved to Cambridge, to professor Magdalena Zernicka-Goetz’s group at the Wellcome Trust/Cancer Research UK Gurdon Institute, where I have just completed my PhD. Here, I am investigating the molecular and cellular mechanisms behind the first cell fate decision that generates pluripotent ICM and the differentiated extra-embryonic lineage, the trophectoderm.
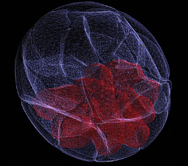
My image shows a three-dimensional reconstruction of a mouse blastocyst. Blastocyst is an outcome of a pre-implantation development, a unique developmental phase characteristic for placental mammals such as mouse or human. In mouse, this phase encompasses a period of approximately 4.5 days and leads to formation of the first tissue types: outer epithelium, called the trophectoderm (TE; in white) and pluripotent group of inner cells, the inner cell mass (ICM; in red). Following implantation into the uterus wall, the ICM will form the fetus, differentiating into all tissue types of the body. TE, on the other hand, will give rise to extra-embryonic structures that support embryo development by mediating nutrients exchange between mother and the fetus and providing signals to pattern the embryo and segregate germ cell lineage. Understanding how these first tissue types emerge during early development becomes increasingly important in modern world, given growing interest in assisted reproductive technology (ART) and associated with it the need for optimizing culture conditions and assessing quality of the obtained embryos.
This image was produced by scanning the embryo with a confocal microscope to create multiple virtual sections, which were then reconstructed using 3D computer software.
I submitted it to the Wellcome Image Awards because I believe it is important that the general public realizes that the pre-implantation embryo is not just a group of pluripotent cells that will build the body of the future individual. In fact, approximately two-thirds of embryos’ cells at that stage are differentiated (the trophectoderm) and will build the extra-embryonic structures. It is important to realize that, given the fierce discussion on the moral aspects of the research on the embryo derived ESc.”
 Kara Cerveny
Kara Cerveny
“I’m a cell and developmental biologist with an interest in the developing nervous system. Up until a few months ago, I was a post-doc in Steve Wilson’s lab at the University College London where I studied the transition from proliferation to differentiation in the zebrafish retina.
This image highlights two separate populations within the zebrafish retinal stem cell zone, an area found in the region of the retina closest to the lens. The undifferentiated retinal stem cells are highlighted in red, while the cells that are beginning to differentiate are highlighted in purple. The central yellow region is the lens.
I took this image relatively early in my post-doc (nearly 5 years ago now) when I was working out the protocol of double in situ staining for fish eyes. This particular sample worked beautifully, and to be a bit creative and make a pretty picture to hang next to my computer, I cropped the original image so that only the top half was visible and then reflected it across the absissa to make the image you now see. Eventually, unmodified images similar to this one were used for a paper that was published in Development.
When I presented my work at the Fall 2010 BSDB sensory biology meeting last September, Laura Pastorelli, the image curator for the Wellcome Trust, asked me if I would be willing to submit this image to the Wellcome Image collection. I was surprised and very happy when I learned that my image had been chosen to be part of this year’s award series. I later learned that the judges had been captivated by the kaleidoscope effect created by the elongated retinal progenitor cells seeming to radiate from the lens (just like I had been several years before).”
 (4 votes)
(4 votes)
 Loading...
Loading...


 (1 votes)
(1 votes)





