The people behind the papers: Matthias Tisler & Martin Blum
Posted by the Node Interviews, on 21 February 2017
Conjoined twins have fascinated biologists for centuries. In twins joined at the thorax, left-right patterning is disrupted, but only in one half of the right hand twins. Today’s paper, from this week’s issue of Current Biology, tackles this enigmatic phenomenon using Xenopus, and reveals that laterality in conjoined twins is determined by cilia-driven leftward flow. We caught up with lead author and soon-to-graduate PhD student Matthias Tisler, and his PI Martin Blum of Hohenheim University in Germany.

So Martin, can tell us your scientific biography and the aims of the Blum lab?
MB Well, it took some time until I became an embryologist. I trained as a biologist and got my Ph.D. in Karlsruhe, Germany, working with Peter Herrlich on DNA repair and trying to clone the gene defective in patients with Xeroderma pigmentosum. A first postdoc led me to the Biocenter in Basel, where we studied genetic polymorphisms of drug metabolism in the lab of Urs Mayer and cloned the genes and variants responsible for slow acetylation. This is where I met Walter Gehring, whose work I had followed and admired for a very long time. He encouraged me to switch to developmental biology even at a relatively advanced stage of my career, and he introduced me to Eddy De Robertis, with whom I did a second postdoc in LA.
These were exciting years: the lab was at the forefront of unraveling the molecular secrets of Spemann’s organizer and cloned some of the first genes, such as Goosecoid and Chordin. After my return to Germany I set up my own lab and serendipitously cloned Pitx2, in an attempt to clone Gsc-related genes. From that time onwards, my lab has studied left-right asymmetry in various organisms, mouse and frog but also rabbit, pig and even sea urchins. We try to understand why and how left-right organ asymmetry evolved, at what stage cilia became instrumental, and how cilia-driven symmetry breakage works.
What is research like for developmental biology in Stuttgart?
MB Hohenheim is a small university and there is just one other embryology lab working on Drosophila. However, we are very close to Tübingen and Heidelberg, and we have established close ties with other Xenopus labs all over Germany. Therefore, we are not isolated at all, and we have quite a good standing with biology students at all levels that are interested in joining the lab for summer projects or their bachelor or master thesis. The lab usually consists of 5-6 Ph.D. students, 2-3 technicians, a staff scientist and numerous bachelor and master students. We also have two junior groups in the institute, with whom we have lab meetings and seminars together. I love this place, the beautiful campus and the small size which allows you to basically know all your colleagues and which makes for a very pleasant atmosphere.
And Matthias, how did you come to join Martin’s lab?
MT Back in 2006, when I started my studies in Hohenheim, Martin was in charge of introducing molecular cell biology and in the course of his lectures at one point passed a cartilage-stained E14.5 mouse embryo around. I think it was on one of my very first days at Hohenheim University. After this lecture, I was completely fascinated by the beauty and complexity of developmental biology and wanted to join Martin’s lab as soon as possible. After some time, I finally had the courage to ask for a job in the lab, just to get in contact with the people and to get my hands on embryos. Martin gave me a chance…and I think I did well over the years.
What can conjoined twins tell us about development?
MT Dating back to the experiments of Hans Spemann and colleagues, twins and their experimental induction led not only to the discovery of the famous Spemann organizer but also led him to think about the establishment of left-right asymmetry. So twinning has a long tradition in inspiring developmental biology.
MB I couldn’t agree more: conjoined twins have been a great assay ever since Hilde Mangold’s organizer transplantation in newt embryos in the 1920s in Hans Spemann’s laboratory that induced the formation of a conjoined twin on the ventral side. The molecular elucidation of the organizer used this assay time and again to demonstrate the potential of genes such as Wnt8 or cerberus to elicit the organizer phenomenon. For example, many Wnt pathway components have been investigated and their epistatic relationships unravelled in that assay, i.e. the dependence of twinning on the presence of such factors.
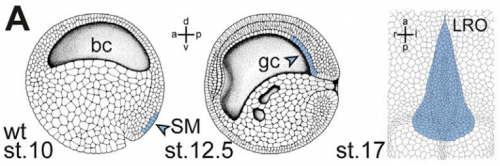
And why are Xenopus in particular a good model for twinning and laterality?
MT Xenopus is THE ideal model organism to induce a secondary body axis by the (even sided) injections of mRNAs during early stages of development. As frog embryos develop in a petri-dish, they are suitable for additional manipulations like morpholino oligomer-mediated gene knockdowns or – in experiments that we have used in the present study – injection of methylcellulose (wallpaper paste) into the archenteron to block cilia-driven leftward flow. And: all these treatments can be combined, that is only possible in Xenopus embryos.
MB By these sided injections, which Matthias mentioned, one can assess whether a gene works on the left or right side, and the contralateral (uninjected) side always serves as an internal control.
Can you give us the key results of the paper in a paragraph?
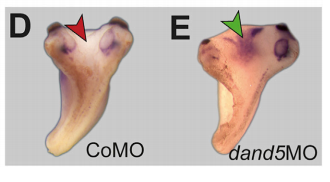
MB & MT The observation that the heart loops normally in left conjoined twins but is randomized in right twins (50% normal, 50% inverted) is old and has been reported in numerous human cases. Spemann reproduced this finding by performing partial ligatures of early stage newt and frog embryos. We injected ß-catenin to induce twinning at will on the left or right side of the endogenous twin and observed the same phenomenon. While two clearly and completely separated precursor tissues of the ciliated left-right organizer (LRO) formed in all cases, the LRO was partially fused, i.e. the right margin of the LRO in the left twin was joined to the left part of the LRO of the right one. Cilia were present on both sides, of normal length and polarity, and they were motile and produced a leftward fluid flow on both sides. Yet the asymmetric Nodal signaling cascade, which determines heart looping and the placement of the other asymmetric organs in the chest and abdomen, was only induced in the left but not in the right twin. This was consistent with the observed heart situs (normal in left and random in right twins), but odd, as flow was normal also in the right twin. A possible solution surfaced when we analyzed the Nodal inhibitor Dand5, which is coexpressed with Nodal at the LRO margins on both sides: in conjoined right twins, flow down-regulates Dand5 like in left ones, but only partially, likely because there is a vast excess available from the fused right side of the left twin. This observation predicted that a knockdown of Dand5 on the right side of the left twin should induce the asymmetric gene cascade also in the right twin, which is exactly what we observed in an experiment, that can only be performed in Xenopus. To round this off, we manipulated Dand5 and flow (using methylcellulose) in a sided manner and were able to induce the Nodal cascade in twins at will, which demonstrated that flow and its target Dand5 also determine laterality in twins, just as in singletons.
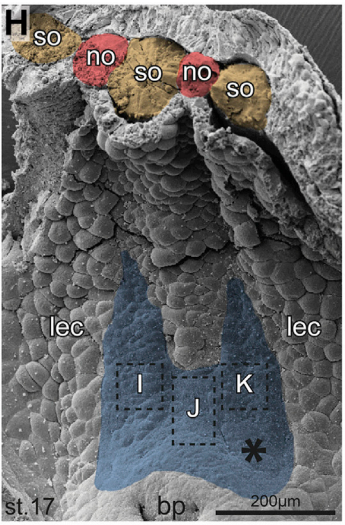
And your results overcome any final objections to the role of cilia in breaking symmetry?
MB & MT A role for cilia-mediated symmetry breakage in fish, amphibian and mammalian embryos has been demonstrated in many genetic and embryological studies. However, it has been argued that cilia-driven leftward flow merely amplifies an earlier asymmetry which is present already during early cleavage stages (flow only sets in during neurulation). This argument was mostly based on organ situs determination in conjoined twins, which is triggered by activation of the Spemann organizer on the ventral side. Because the organizer acts during gastrulation, it was assumed that cilia could not directly impact on laterality determination. Our work now shows that the initially clearly separated axes fuse in a way that flow in the right twin is insufficient to completely repress the Nodal inhibitor Dand5. Because this riddle now is solved, there are no experiments left that argue against cilia and flow.
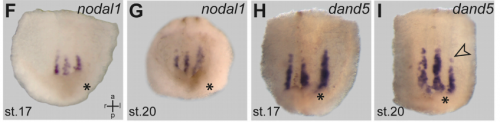
Is the same process is occurring in conjoined humans?
MB & MT We are pretty sure that this mechanism also works in humans: the twins that we can generate in Xenopus are fused at the thorax and have two heads, which is exactly the type of human twins that show this odd laterality defects (and which constitute some 70% of human conjoined twins. Also, the anterior-posterior position of the LRO corresponds to the thorax in the adult. Human twins fused at other sites show mostly normal organ situs, for example if they are just fused at the heads. In these cases, the LROs would be expected to be completely separated during neurulation.
When doing the research, was there a particularly exciting result or eureka moment that has stayed with you?
MT The initial observation of the duplication of the Left-Right Organizer in twins and that this could be the key to explain the century-old question of how the organ situs is determined in conjoined twins will stay with me.
What about the flipside: any particular moments of frustration or despair?
MT Some moments of frustration that probably everybody working with frog embryos shares is when the manipulated specimens do not survive the night. Specifically regarding the twin project: the task of visualizing the flow in the archenteron of conjoined twins has had a lot of “potential” to causing frustration and despair.
And what next for you Matthias?
MT Now that my time in Hohenheim is ending (the defense of my thesis has been scheduled), it is time for me to try something new. As a consequence of this work I really got interested in human development and disease, so I decided to switch gears and to go to Med School. In the future I hope to be working as a clinical scientist from bench to bedside and backwards.
And where next for the Blum lab?
MB Off to the next frontier: linking the flow-dependent repression of Dand5 to upstream events. We really need to understand how this repression is brought about in a flow- and cilia-dependent manner. This task has the potential to keep us busy for some time. And most likely we will be using the twin assay again.
Matthias Tisler, Thomas Thumberger, Isabelle Schneider, Axel Schweickert & Martin Blum. 2017. Leftward Flow Determines Laterality in Conjoined Twins. Current Biology, 27 (4): 543–548.
Browse the People Behind the Papers archive here.


 (3 votes)
(3 votes)
 (No Ratings Yet)
(No Ratings Yet)




 This report written by Justine Alford and highlighting a recent Development paper originally appeared on the
This report written by Justine Alford and highlighting a recent Development paper originally appeared on the 



 Auxin – a key plant hormone – plays a prominent role in regulating plant developmental processes, and delineating its role is therefore the subject of intensive investigation. In their
Auxin – a key plant hormone – plays a prominent role in regulating plant developmental processes, and delineating its role is therefore the subject of intensive investigation. In their  Cellular metabolism plays a vital role in development, beyond the simple production of energy, and may be involved in the regulation of cell fate. In their
Cellular metabolism plays a vital role in development, beyond the simple production of energy, and may be involved in the regulation of cell fate. In their  The process of neural tube closure is complex and involves cellular events such as convergent extension, apical constriction and interkinetic nuclear migration, as well as precise molecular control via the non-canonical Wnt/planar cell polarity pathway, Shh/BMP signalling, and the transcription factors Grhl2/3, Pax3, Cdx2 and Zic2. More recently, biomechanical inputs into neural tube morphogenesis have also been identified. In their
The process of neural tube closure is complex and involves cellular events such as convergent extension, apical constriction and interkinetic nuclear migration, as well as precise molecular control via the non-canonical Wnt/planar cell polarity pathway, Shh/BMP signalling, and the transcription factors Grhl2/3, Pax3, Cdx2 and Zic2. More recently, biomechanical inputs into neural tube morphogenesis have also been identified. In their