Jens Lüders leads the Microtubule Organization laboratory (Photo: Battista/Minocri, IRB Barcelona)

“la Caixa” PhD student Nicolas Lecland is the first author of the study published in Nature Cell Biology (Photo: Battista/Minocri, IRB Barcelona)
A breakthrough at IRB Barcelona fills a knowledge gap in understanding how the cell division apparatus, the mitotic spindle, is formed.
The in vivo visualization and monitoring of the starting points of microtubules — filaments responsible for organising the mitotic spindle — provides novel insight into the dynamic architecture of this structure.
The findings will also contribute to understanding how the mitotic spindle is perturbed by drugs that target microtubules and that are used in chemotherapy.
The division of a cell in two requires the assembly of the mitotic spindle, an extremely complex structure, which is the result of the coordinated action of a multitude of proteins and a finely tuned balance of their activities. A large part of the time that a cell requires to divide is devoted to assembling the mitotic spindle, which, superficially, resembles a ball of thread with the shape of a rugby ball.
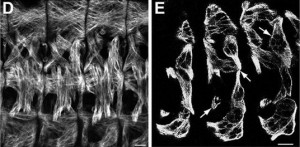
The most abundant components of the spindle are the microtubules. “By labelling the ends of thousands of these fine filaments, which are indispensable and extremely dynamic and variable, we have finally been able to follow their distribution and movement during the assembly of the mitotic spindle,” explains Jens Lüders, a cell biologist from the Institute for Research in Biomedicine (IRB Barcelona). The breakthrough appeared yesterday in the advanced online edition of the journal Nature Cell Biology.
“For more than 10 years we have been able to track only the growing ends of microtubules but not the starting points. As a result, we lacked essential information in order to understand the dynamic architecture of the mitotic spindle and how it contributes to cell division,” says Lüders. Headed by the German scientist who runs the Microtubule Organisation group at IRB Barcelona, the study carries only two names, his own and that of the French researcher Nicolas Lecland, first author, who completed his PhD at IRB Barcelona through a “la Caixa” fellowship.
The scientists have demonstrated that the protein γ-tubulin localizes at the starting points of the microtubule filaments and is relatively stably associated with these structures. Using a version of γ-tubulin that carries a fluorescent label activated by laser light, the researchers were able to follow the movement of the starting points of microtubules within mitotic spindles by filming dividing human cells.
The Advanced Digital Microscopy Facility, a joint IRB Barcelona-Barcelona Science Park Facility run by the IRB physicist Julien Colombelli, has been crucial for setting up the technology required. “The success of this study is also the result of the technical know-how and cutting-edge technology available, without which we would never have been able to tackle this project,” emphasizes Lüders.
The researchers describe for the first time where most microtubules form inside the mitotic spindle, how they develop, and how their starting points are transported—with the help of three motor proteins—to opposite poles of the spindle, where they attach. Simultaneous to this process, the opposite ends of the filaments extend towards the cell centre, where they interact with chromosomes.
When the spindle is finally assembled, the microtubules pull the chromosomes to opposite poles and initiate the physical division of the cell. “We now have a more complete understanding of how the spindle assembles and functions and can use our novel marker for testing old and new hypotheses about underlying mechanisms,” says the scientist.
A new tool to study cancer
In addition, the breakthrough paves the way to “better” understanding the mode of action of drugs that inhibit microtubules and that are used in chemotherapy. These kinds of drugs impede the mitotic spindle, thus preventing cell division and interfering with tumour growth.
In spite of the many years of clinical success of these treatments against cancer, little is known about how they impair spindle architecture and function. Although these drugs are highly efficient, they do not show the specificity desirable as they also affect healthy dividing cells. In addition, they affect non-dividing cells such as neurons, in which microtubules also have important functions.
“A better understanding of the differences in spindle organisation between cancer and healthy cells and how they respond to microtubule-targeted drugs is essential in order to optimise treatments, for example by identifying more specific drugs or new targets. This tool could be useful to achieve these objectives,” states the researcher.
The study has been supported by structural funds from the Generalitat de Catalunya, a Marie Curie grant from the European Union, and the Plan Nacional, of the Ministry of Economy and Competitiveness.
Reference article:
The dynamics of microtubule minus ends in the human mitotic spindle
Nicolas Lecland and Jens Lüders
Nature Cell Biology (2014) Doi: http://dx.doi.org/10.1038/ncb2996
Video: gtubpaGFPmerge copy
This article was first published on the 30th of June 2014 in the news section of the IRB Barcelona website
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (1 votes)
(1 votes)


 (No Ratings Yet)
(No Ratings Yet) Embryonic stem cell (ESC) cultures display a marked heterogeneity in the expression of Nanog, one of several core pluripotency factors required for proper development in vivo. In addition, Nanog levels have also been shown to fluctuate in individual ESCs in vitro; however, the extent and functional consequences of these fluctuations in different pluripotency states has not been fully established. Now, on p.
Embryonic stem cell (ESC) cultures display a marked heterogeneity in the expression of Nanog, one of several core pluripotency factors required for proper development in vivo. In addition, Nanog levels have also been shown to fluctuate in individual ESCs in vitro; however, the extent and functional consequences of these fluctuations in different pluripotency states has not been fully established. Now, on p.  The balance between excitatory versus inhibitory neuron specification during development is crucial for sensory information processing in later life. The basic helix-loop-helix (bHLH) transcription factors Ascl1 and Ptf1a are crucial for establishing this specificity in the dorsal spinal cord, but how these two factors, which recognise a similiar DNA motif, can have opposite downstream effects is unclear. Now, on p.
The balance between excitatory versus inhibitory neuron specification during development is crucial for sensory information processing in later life. The basic helix-loop-helix (bHLH) transcription factors Ascl1 and Ptf1a are crucial for establishing this specificity in the dorsal spinal cord, but how these two factors, which recognise a similiar DNA motif, can have opposite downstream effects is unclear. Now, on p.  During blastocyst development, asymmetric cell divisions generate polar and apolar daughter cells, which organise into outer and inner positions, respectively, to form the trophectoderm (TE) and inner cell mass (ICM) lineages. The Hippo signaling pathway is crucial for setting up this early lineage specification, but how Hippo signaling relates to cell position and polarity remains unclear. In this issue (p.
During blastocyst development, asymmetric cell divisions generate polar and apolar daughter cells, which organise into outer and inner positions, respectively, to form the trophectoderm (TE) and inner cell mass (ICM) lineages. The Hippo signaling pathway is crucial for setting up this early lineage specification, but how Hippo signaling relates to cell position and polarity remains unclear. In this issue (p.  Cell polarity is fundamental for biological activity across many varied cell types within different animal species. Intracellular trafficking regulates the differential distribution of proteins that is fundamental to establishing cell polarity, but how cell polarity regulators exert their effects on trafficking machinery is largely unknown. Now, on p.
Cell polarity is fundamental for biological activity across many varied cell types within different animal species. Intracellular trafficking regulates the differential distribution of proteins that is fundamental to establishing cell polarity, but how cell polarity regulators exert their effects on trafficking machinery is largely unknown. Now, on p.  In March 2014, the RIKEN Center for Developmental Biology in Kobe, Japan, hosted a meeting entitled ‘Regeneration of Organs: Programming and Self-Organization’. Scientists from across the globe met to discuss current research on regeneration, organ morphogenesis and self-organization – and the links between these fields. As discussed by Daniel Goldman, a diverse range of experimental models and organ systems was presented at the meeting, and the speakers aptly illustrated the unique power of each. See the Meeting Review on p
In March 2014, the RIKEN Center for Developmental Biology in Kobe, Japan, hosted a meeting entitled ‘Regeneration of Organs: Programming and Self-Organization’. Scientists from across the globe met to discuss current research on regeneration, organ morphogenesis and self-organization – and the links between these fields. As discussed by Daniel Goldman, a diverse range of experimental models and organ systems was presented at the meeting, and the speakers aptly illustrated the unique power of each. See the Meeting Review on p  Branching morphogenesis is the developmental program that builds the epithelial trees of various organs, including the airways of the lung, the collecting ducts of the kidney, and the ducts of the mammary and salivary glands. R
Branching morphogenesis is the developmental program that builds the epithelial trees of various organs, including the airways of the lung, the collecting ducts of the kidney, and the ducts of the mammary and salivary glands. R The formation of the vasculature is essential for tissue maintenance and regeneration, and understanding how vascular formation is coordinated in vivo can offer valuable insights into engineering approaches for therapeutic vascularization and angiogenesis. Here, Kyung Min Park and Sharon Gerecht discuss how the process of vascular development can be used to guide approaches to engineering vasculature. See the Review on p.
The formation of the vasculature is essential for tissue maintenance and regeneration, and understanding how vascular formation is coordinated in vivo can offer valuable insights into engineering approaches for therapeutic vascularization and angiogenesis. Here, Kyung Min Park and Sharon Gerecht discuss how the process of vascular development can be used to guide approaches to engineering vasculature. See the Review on p.