In Development this week (Vol. 139, Issue 12)
Posted by Seema Grewal, on 22 May 2012
Here are the highlights from the current issue of Development:
Mechanical changes in cochlea development
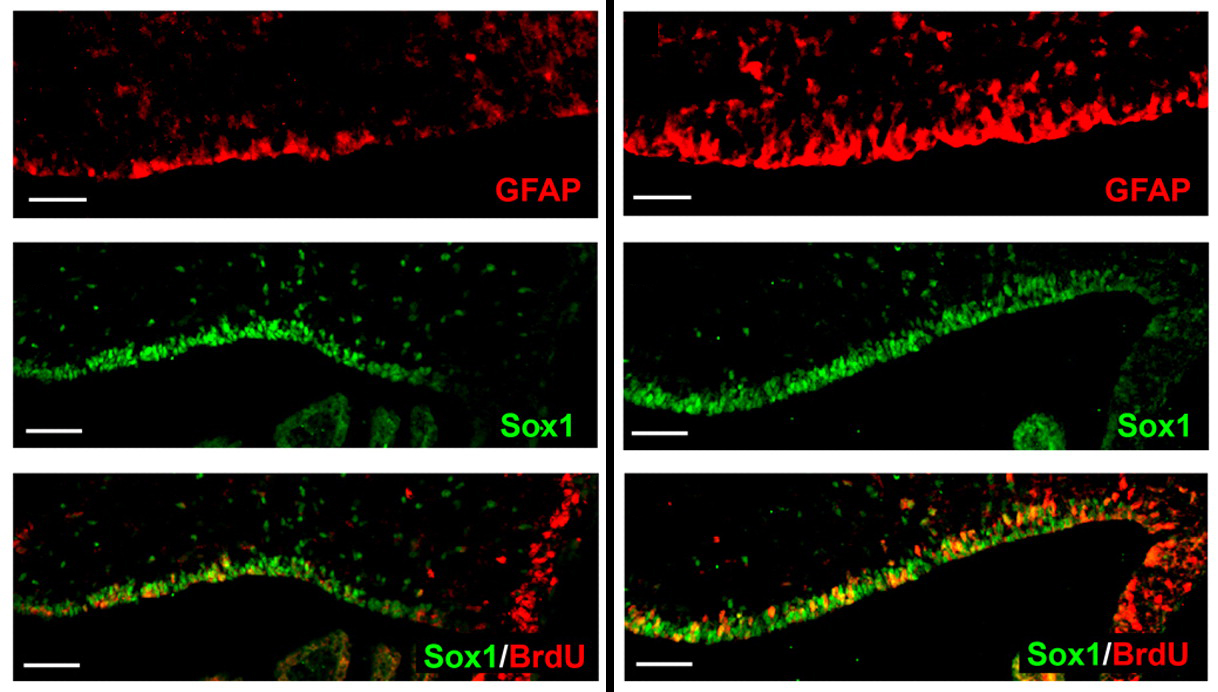
 Correct patterning of the mammalian inner ear sensory epithelium, which contains mechanosensory outer hair cells (OHCs) that detect and amplify sound vibrations and non-sensory supporting cells such as pillar cells (PCs), is essential for hearing. The cell surface mechanical properties of both OHCs and PCs are important for their function but how are these properties regulated during development? On p. 2187, Katherine Szarama and colleagues use atomic force microscopy to show that OHCs and PCs have different cell surface mechanical properties that develop over different time courses. By pharmacologically modulating cytoskeletal elements, they show that the increase in OHC stiffness observed during development depends primarily on actin whereas the development of the cell surface mechanical properties of PCs depends on microtubules. In addition, they report that fibroblast growth factor signalling regulates the developing cell surface mechanical properties of OHCs and PCs, in part by altering cytoskeletal dynamics. These new insights into inner ear development may eventually lead to better treatments for hearing loss.
Correct patterning of the mammalian inner ear sensory epithelium, which contains mechanosensory outer hair cells (OHCs) that detect and amplify sound vibrations and non-sensory supporting cells such as pillar cells (PCs), is essential for hearing. The cell surface mechanical properties of both OHCs and PCs are important for their function but how are these properties regulated during development? On p. 2187, Katherine Szarama and colleagues use atomic force microscopy to show that OHCs and PCs have different cell surface mechanical properties that develop over different time courses. By pharmacologically modulating cytoskeletal elements, they show that the increase in OHC stiffness observed during development depends primarily on actin whereas the development of the cell surface mechanical properties of PCs depends on microtubules. In addition, they report that fibroblast growth factor signalling regulates the developing cell surface mechanical properties of OHCs and PCs, in part by altering cytoskeletal dynamics. These new insights into inner ear development may eventually lead to better treatments for hearing loss.
Resetting after quiescence
 During development, networks of regulatory genes control precisely timed sequences of developmental events. In C. elegans, heterochronic genes, which encode several transcription factors and microRNAs (miRNAs) that regulate the expression of these transcription factors, control stage-specific cell-fate decisions. Under adverse conditions, however, second larval stage (L2) worms enter a quiescent state called dauer. Intriguingly, when conditions improve, dauer larvae complete development normally. Here (p. 2177), Xantha Karp and Victor Ambros investigate how cell-fate progression is reset after dauer. Progression from L2 to L3 requires downregulation of the transcription factor Hunchback-like-1 (HBL-1), and, during continuous development, HBL-1 downregulation relies mainly on three let-7 family miRNAs. However, after dauer, the researchers report, lin-4 miRNA and an altered set of let-7 family miRNAs downregulate HBL-1. This shift in the programming of HBL-1 downregulation, they propose, involves the enhancement of lin-4 and let-7 miRNA activity by miRNA-induced silencing complex (miRISC) modulators. The employment of alternative genetic regulatory pathways can, therefore, ensure the robust progression of cell-fate specification after temporary developmental quiescence.
During development, networks of regulatory genes control precisely timed sequences of developmental events. In C. elegans, heterochronic genes, which encode several transcription factors and microRNAs (miRNAs) that regulate the expression of these transcription factors, control stage-specific cell-fate decisions. Under adverse conditions, however, second larval stage (L2) worms enter a quiescent state called dauer. Intriguingly, when conditions improve, dauer larvae complete development normally. Here (p. 2177), Xantha Karp and Victor Ambros investigate how cell-fate progression is reset after dauer. Progression from L2 to L3 requires downregulation of the transcription factor Hunchback-like-1 (HBL-1), and, during continuous development, HBL-1 downregulation relies mainly on three let-7 family miRNAs. However, after dauer, the researchers report, lin-4 miRNA and an altered set of let-7 family miRNAs downregulate HBL-1. This shift in the programming of HBL-1 downregulation, they propose, involves the enhancement of lin-4 and let-7 miRNA activity by miRNA-induced silencing complex (miRISC) modulators. The employment of alternative genetic regulatory pathways can, therefore, ensure the robust progression of cell-fate specification after temporary developmental quiescence.
ExE progenitors make an eXit
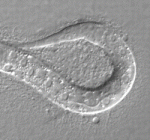
 In female mammalian embryos, inactivation of one of the two X chromosomes in each cell regulates X-linked gene expression. X chromosome inactivation (XCI) is dependent on the non-coding RNA Xist, which is expressed from and coats the inactivated X chromosome. Inheritance of a paternally derived Xist mutation causes embryonic lethality because the inactivation of the paternally inherited X chromosome that occurs in the extra-embryonic lineages of female mouse embryos during imprinted XCI fails. Now, Terry Magnuson and colleagues (p. 2130) describe the exact consequences of failed XCI within the extra-embryonic ectoderm (ExE). The ExE of X/XXist– embryos consists mainly of differentiated giant cells and their progenitors, they report, and less differentiated spongiotrophoblast precursors are not maintained. At E6.5, the ExE lacks CDX2, which is required to maintain the ExE’s multipotent state. Moreover, trophoblast stem cell lines derived from X/XXist– blastocysts completely reverse normal imprinted XCI patterns. These results suggest that dosage compensation is indispensable for the maintenance of trophoblast progenitors and that imprinted XCI is probably erased in ExE cells.
In female mammalian embryos, inactivation of one of the two X chromosomes in each cell regulates X-linked gene expression. X chromosome inactivation (XCI) is dependent on the non-coding RNA Xist, which is expressed from and coats the inactivated X chromosome. Inheritance of a paternally derived Xist mutation causes embryonic lethality because the inactivation of the paternally inherited X chromosome that occurs in the extra-embryonic lineages of female mouse embryos during imprinted XCI fails. Now, Terry Magnuson and colleagues (p. 2130) describe the exact consequences of failed XCI within the extra-embryonic ectoderm (ExE). The ExE of X/XXist– embryos consists mainly of differentiated giant cells and their progenitors, they report, and less differentiated spongiotrophoblast precursors are not maintained. At E6.5, the ExE lacks CDX2, which is required to maintain the ExE’s multipotent state. Moreover, trophoblast stem cell lines derived from X/XXist– blastocysts completely reverse normal imprinted XCI patterns. These results suggest that dosage compensation is indispensable for the maintenance of trophoblast progenitors and that imprinted XCI is probably erased in ExE cells.
Gibberellin regulation of flowering
 Several environmental cues, including day length, and endogenous developmental signals regulate the transition from leaf production to flower formation in plants. In Arabidopsis, the growth regulator gibberellin promotes this transition most strongly under short day (SD) conditions. Here (p. 2198), George Coupland and colleagues show how gibberellins also promote flowering in response to long days (LDs). The researchers deplete gibberellins in the vascular tissue or the shoot apical meristem by tissue-specific overexpression of GA2ox7, which catabolises gibberellins. Under LD conditions, gibberellins are needed in the vascular tissue to increase production of a systemic signal that is transported from the leaves to the meristem during floral induction. However, in the meristem, instead of activating the expression of the transcription factor SOC1 (which is needed to induce flowering under SD conditions), in response to LDs, gibberellins regulate the expression of SPL transcription factors, which are needed later during floral induction. Thus, the researchers conclude, gibberellins play spatially distinct roles in promoting flowering under long photoperiods.
Several environmental cues, including day length, and endogenous developmental signals regulate the transition from leaf production to flower formation in plants. In Arabidopsis, the growth regulator gibberellin promotes this transition most strongly under short day (SD) conditions. Here (p. 2198), George Coupland and colleagues show how gibberellins also promote flowering in response to long days (LDs). The researchers deplete gibberellins in the vascular tissue or the shoot apical meristem by tissue-specific overexpression of GA2ox7, which catabolises gibberellins. Under LD conditions, gibberellins are needed in the vascular tissue to increase production of a systemic signal that is transported from the leaves to the meristem during floral induction. However, in the meristem, instead of activating the expression of the transcription factor SOC1 (which is needed to induce flowering under SD conditions), in response to LDs, gibberellins regulate the expression of SPL transcription factors, which are needed later during floral induction. Thus, the researchers conclude, gibberellins play spatially distinct roles in promoting flowering under long photoperiods.
Migrating primordial germ cells exploit endoderm remodelling
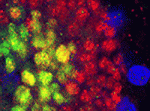 Cell migration through epithelial tissues occurs during development, infection, inflammation, wound healing and cancer metastasis. But how do cells overcome the impermeable junctions between epithelial cells? Leukocytes move out of blood vessels by loosening endothelial cell-cell junctions but do all cells actively remodel tissue barriers during migration? According to Jessica Seifert and Ruth Lehmann, who are studying Drosophila primordial germ cell (PGC) migration through the endodermal epithelium to the gonadal mesoderm, the answer to this question is no (see p. 2101). Although PGC migration requires activation of the G protein-coupled receptor Trapped in endoderm 1 (Tre1) within PGCs, the timing of PGC migration is dictated by the developmental stage of the endoderm. Now, using live imaging and genetic manipulation, the researchers show that PGCs take advantage of developmentally regulated epithelial remodelling, which causes discontinuities in the endoderm, to gain access to the gonadal mesoderm. Thus, Seifert and Lehmann conclude that, rather than actively remodelling tissue barriers, some migrating cells exploit existing tissue permeability.
Cell migration through epithelial tissues occurs during development, infection, inflammation, wound healing and cancer metastasis. But how do cells overcome the impermeable junctions between epithelial cells? Leukocytes move out of blood vessels by loosening endothelial cell-cell junctions but do all cells actively remodel tissue barriers during migration? According to Jessica Seifert and Ruth Lehmann, who are studying Drosophila primordial germ cell (PGC) migration through the endodermal epithelium to the gonadal mesoderm, the answer to this question is no (see p. 2101). Although PGC migration requires activation of the G protein-coupled receptor Trapped in endoderm 1 (Tre1) within PGCs, the timing of PGC migration is dictated by the developmental stage of the endoderm. Now, using live imaging and genetic manipulation, the researchers show that PGCs take advantage of developmentally regulated epithelial remodelling, which causes discontinuities in the endoderm, to gain access to the gonadal mesoderm. Thus, Seifert and Lehmann conclude that, rather than actively remodelling tissue barriers, some migrating cells exploit existing tissue permeability.
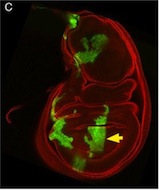
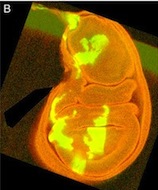
Tcf21 seals cardiac fibroblast fate
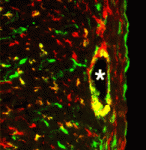 The primary source of cardiac fibroblasts, which are essential for normal heart physiology, is a subpopulation of epicardial cells that has undergone epithelial-to-mesenchymal transition (EMT) and entered the myocardium. But does cardiac fibroblast specification occur early in the formation of the epicardium (which is a multipotent mesothelial layer of cells that spreads over the developing myocardium) or after the epicardial-derived cells have entered the myocardium? Here (p. 2139), Michelle Tallquist and colleagues resolve this puzzle by investigating the role of the transcription factor Tcf21 in cardiac fibroblast specification in mice. The researchers use lineage tracing to show that Tcf21-expressing epicardial cells are committed to the cardiac fibroblast lineage before the initiation of epicardial EMT. Moreover, Tcf21-null embryos fail to develop cardiac fibroblasts and Tcf21-null fibroblast progenitors do not undergo EMT. These results indicate that cardiac fibroblast specification occurs in the epicardium before EMT occurs and, importantly, these findings identify Tcf21 as an essential transcription factor for cardiac fibroblast cell-fate determination.
The primary source of cardiac fibroblasts, which are essential for normal heart physiology, is a subpopulation of epicardial cells that has undergone epithelial-to-mesenchymal transition (EMT) and entered the myocardium. But does cardiac fibroblast specification occur early in the formation of the epicardium (which is a multipotent mesothelial layer of cells that spreads over the developing myocardium) or after the epicardial-derived cells have entered the myocardium? Here (p. 2139), Michelle Tallquist and colleagues resolve this puzzle by investigating the role of the transcription factor Tcf21 in cardiac fibroblast specification in mice. The researchers use lineage tracing to show that Tcf21-expressing epicardial cells are committed to the cardiac fibroblast lineage before the initiation of epicardial EMT. Moreover, Tcf21-null embryos fail to develop cardiac fibroblasts and Tcf21-null fibroblast progenitors do not undergo EMT. These results indicate that cardiac fibroblast specification occurs in the epicardium before EMT occurs and, importantly, these findings identify Tcf21 as an essential transcription factor for cardiac fibroblast cell-fate determination.
Plus…
X chromosome inactivation in the cycle of life
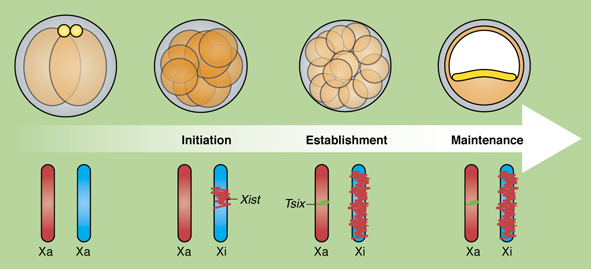 Bakarat and Gribnau review new insights into the molecular events occurring during the life cycle of X chromosome inactivation and, in the accompanying poster, provide an overview of the mechanisms regulating X inactivation and reactivation.
Bakarat and Gribnau review new insights into the molecular events occurring during the life cycle of X chromosome inactivation and, in the accompanying poster, provide an overview of the mechanisms regulating X inactivation and reactivation.
See the Development at a Glance poster article on p. 2085
Evolutionary crossroads in developmental biology: cyclostomes
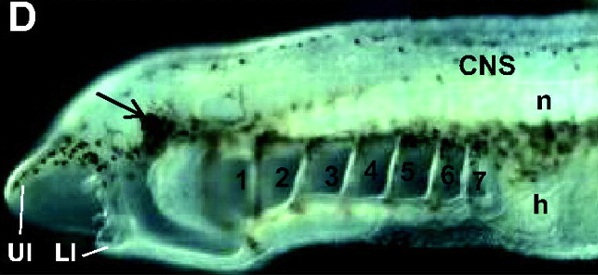 Shimeld and Donoghue summarise the development of cyclostomes (lamprey and hagfish) and discuss how studies of cyclostomes have provided important insight into the evolution of fins, jaws, skeleton and neural crest.
Shimeld and Donoghue summarise the development of cyclostomes (lamprey and hagfish) and discuss how studies of cyclostomes have provided important insight into the evolution of fins, jaws, skeleton and neural crest.
See the Primer article on p. 2091
(note that this article is part of a series of Primer articles on organisms that represent an evolutionary crossroads in the study of evolutionary developmental biology – see the online Featured Topic to view other articles in this series)


 (No Ratings Yet)
(No Ratings Yet)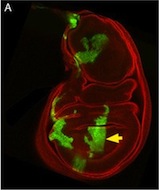
 (4 votes)
(4 votes)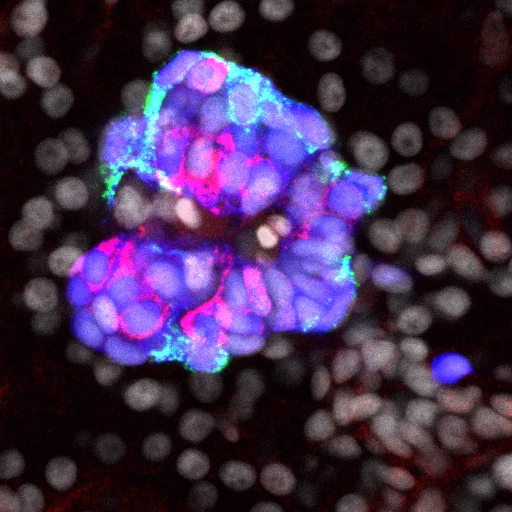
 (6 votes)
(6 votes)