September in preprints
Posted by the Node, on 4 October 2023
Welcome to our monthly trawl for developmental and stem cell biology (and related) preprints.
The preprints this month are hosted on bioRxiv – use these links below to get to the section you want:
- Patterning & signalling
- Morphogenesis & mechanics
- Genes & genomes
- Stem cells, regeneration & disease modelling
- Plant development
- Evo-devo
Developmental biology
| Patterning & signalling
Protein phosphatase 1 regulates core PCP signaling
Song Song, Bomsoo Cho, Alexis T Weiner, Silas Boye Nissen, Irene Ojeda Naharros, Pablo Sanchez Bosch, Kaye Suyama, Yanhui Hu, Li He, Tanya Svinkina, Namrata Udeshi, Steven A Carr, Norbert Perrimon, Jeffrey D. Axelrod
Combinatorial Wnt signaling landscape during brachiopod anteroposterior patterning
Bruno C. Vellutini, José M. Martín-Durán, Aina Børve, Andreas Hejnol
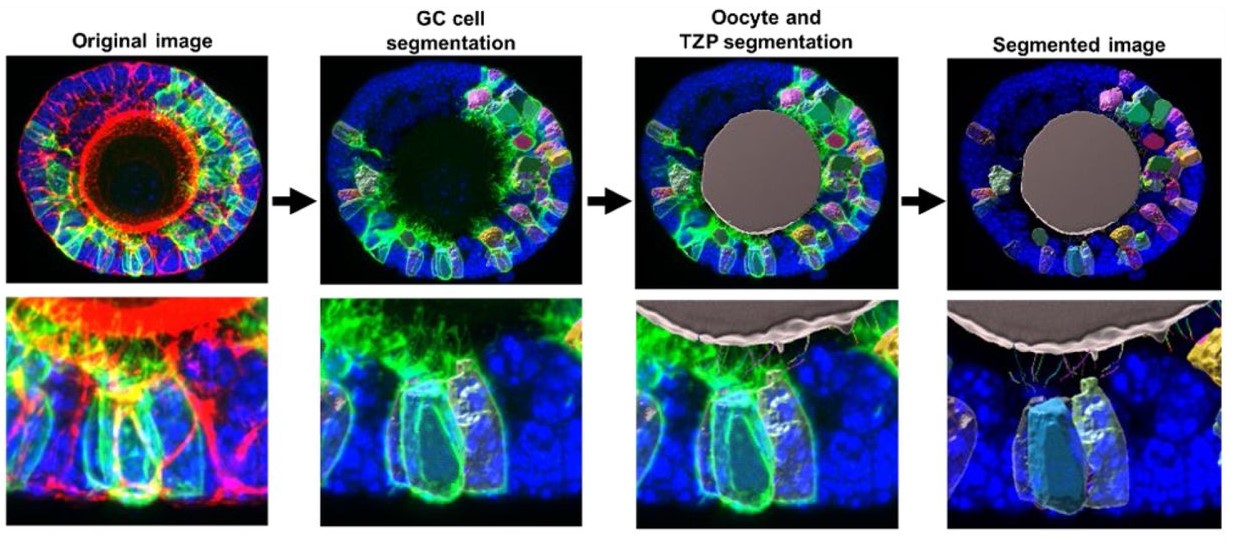
SMAD4 promotes somatic-germline contact during oocyte growth
Sofia Granados-Aparici, Qin Yang, Hugh Clarke
Jianbo Wang, Chenbei Chang, Hwa-seon Seo, Deli Yu, Ivan Popov, Jiahui Tao, Allyson Angermeier, Bingdong Sha, Jeffrey D. Axelrod
Manabu Kitamata, Yoshiaki Otake, Hideaki Kitagori, Xuanshuo Zhang, Yusuke Maki, Rika Boku, Masato Takeuchi, Hideki Nakagoshi
Alonso Rodriguez, Sergio Cordoba, Daniel Delipe-Cordero, Antonio Baonza, David Miguez, Carlos Estella
Marvel Megaly, Gregory Foran, Arsala Ali, Anel Turgambayeva, Ryan D. Hallam, Aleksandar Necakov
Defective mesenchymal Bmpr1a-mediated BMP signaling causes congenital pulmonary cysts
Yongfeng Luo, Ke Cao, Joanne Chiu, Hui Chen, Hong-Jun Wang, Matthew E. Thornton, Brendan H. Grubbs, Martin Kolb, Michael S. Parmacek, Yuji Mishina, Wei Shi
Cell autonomous polarization by the planar cell polarity signaling pathway
Alexis T Weiner, Bomsoo Cho, Kaye Suyama, Jeffrey D Axelrod
Joshua A. Moore, Arielle S. Noah, Eileen W. Singleton, Rosa A. Uribe
Flamingo participates in multiple models of cell competition
Pablo Sanchez Bosch, Bomsoo Cho, Jeffrey D. Axelrod
Matteo A. Negroni, Adria C. LeBoeuf
Miaoxing Wang, Xujun Han, Yunfei Lee, Rie Takayama, Makoto Sato
Embryogenesis in myrmicine ants combines features of short and long germ-band modes of development
Chi-Chun Fang, Arjuna Rajakumar, Andrew Kenny, Ulrich G. Mueller, Ehab Abouheif, David Stein
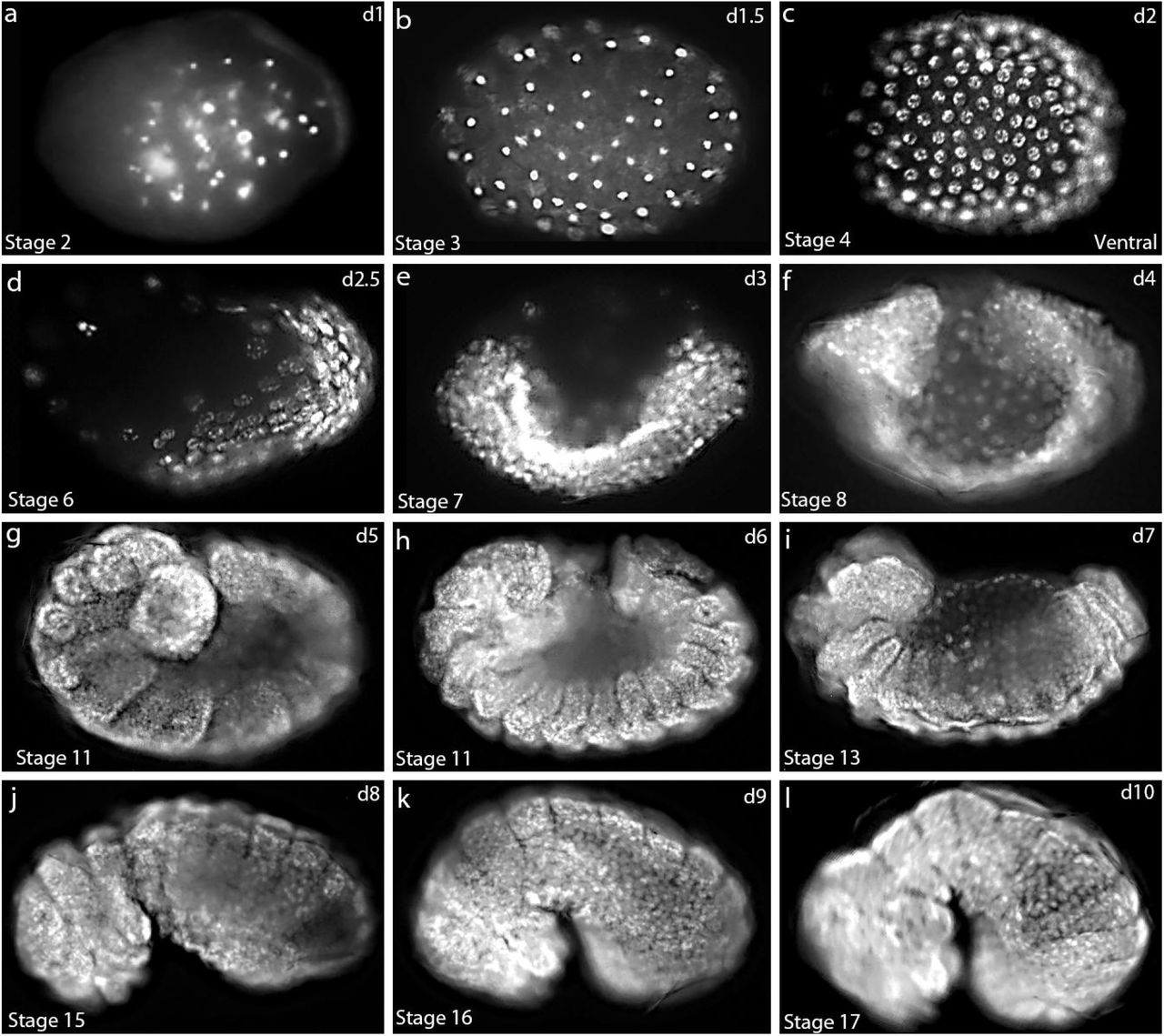
Myosin1G promotes Nodal signaling to control Zebrafish Left-Right asymmetry
Akshai Janardhana Kurup, Florian Bailet, Maximilian Fürthauer
Chhavi Sood, Md Ausrafuggaman Nahid, Kendall R. Branham, Matthew C. Pahl, Susan E. Doyle, Sarah E. Siegrist
| Morphogenesis & mechanics
Vivek Kumar, Prashant Kumar, Takao Hikita, Mingqian Ding, Yukinori Kametani, Masanori Nakayama, Yosuke Hasegawa
Hiroshi Yoke, Atsushi Taniguchi, Shigenori Nonaka
Exploring the principles of embryonic mammary gland branching morphogenesis
Riitta Lindström, Jyoti P. Satta, Satu-Marja Myllymäki, Qiang Lan, Ewelina Trela, Renata Prunskaite-Hyyryläinen, Beata Kaczyńska, Maria Voutilainen, Satu Kuure, Seppo J. Vainio, Marja L. Mikkola
An actomyosin network organizes niche morphology and responds to feedback from recruited stem cells
Bailey N. Warder, Kara A. Nelson, Justin Sui, Lauren Anllo, Stephen DiNardo
Molecular control of cellulosic fin morphogenesis in ascidians
Maxence LANOIZELET, Christel ELKHOURY YOUHANNA, SébasBen DARRAS
Illuminating the Terminal Nerve: Uncovering the Link between GnRH-1 and Olfactory Development
Enrico Amato Jr, Ed Zandro M Taroc, Paolo E. Forni
Keiji Itoh, Olga Ossipova, Miho Matsuda, Sergei Y Sokol
Caitlin C Devitt, Shinuo Weng, Vidal D Bejar-Padilla, José Alvarado, John B Wallingford
Sensitivity of the timing of Drosophila pupal wing morphogenesis to external perturbations
Romina Piscitello-Gómez, Ali Mahmoud, Natalie A Dye, Suzanne Eaton
Overburdened Ferroptotic Stress Impairs Tooth Morphogenesis
H.S. Wang, X.F. Wang, L.Y. Huang, C.L. Wang, F.Y. Yu, L. Ye
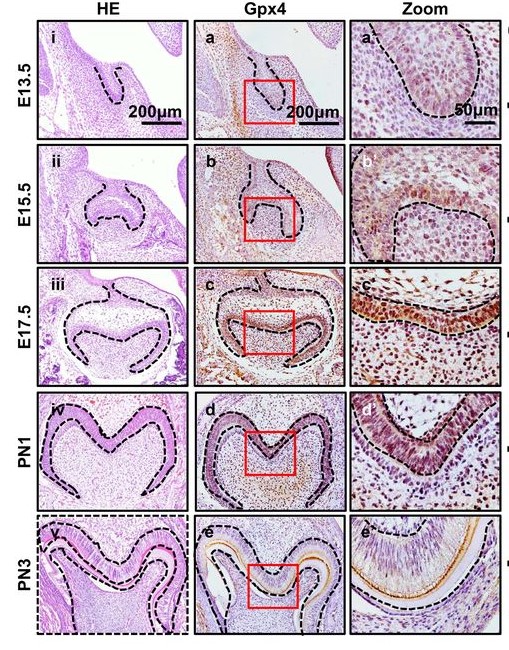
Benjamin Kroeger, Samuel A. Manning, Yoshana Fonseka, Viola Oorschot, Simon A. Crawford, Georg Ramm, Kieran F. Harvey
Elevated temperature fatally disrupts nuclear divisions in the early Drosophila embryo
Girish Kale, Pratika Agarwal, J Jaime Diaz-Larrosa, Steffen Lemke
| Genes & genomes
The autism-associated gene SYNGAP1 regulates human cortical neurogenesis
Marcella Birtele, Ashley Del Dosso, Tiantian Xu, Tuan Nguyen, Brent Wilkinson, Negar Hosseini, Sarah Nguyen, Jean-Paul Urenda, Gavin Knight, Camilo Rojas, Ilse Flores, Alexander Atamian, Roger Moore, Ritin Sharma, Patrick Pirrotte, Randolph S. Ashton, Eric J. Huang, Gavin Rumbaugh, Marcelo P. Coba, Giorgia Quadrato
Xiong Yang, Rong Wan, Zhiwen Liu, Su Feng, Jiaxin Yang, Naihe Jing, Ke Tang
Elizabeth N. Schock, Joshua R York, Austin P Li, Ashlyn Y Tu, Carole LaBonne
ARID1A governs the silencing of sex-linked transcription during male meiosis in the mouse
Debashish U Menon, Prabuddha Chakraborty, Noel Murcia, Terry Magnuson
David Paz, Nayeli G. Reyes-Nava, Briana E. Pinales, Isaiah Perez, Claudia B. Gil, Annalise V. Gonzales, Brian Grajeda, Igor L. Estevao, Cameron C. Ellis, Victoria L. Castro, Anita M. Quintana
Maria M. Mikedis, Bingrun Liu, Dirk G. de Rooij, David C. Page
Mingming Liang, Lichao Zhang, Liangxue Lai, Zhanjun Li
Ada Jimenez-Gonzalez, Federico Ansaloni, Constance Nebendahl, Ghazal Alavioon, David Murray, Weronika Robak, Remo Sanges, Ferenc Müller, Simone Immler
Systematic Perturbation of Thousands of Retroviral LTRs in Mouse Embryos
Jian Yang, Lauryn Cook, Zhiyuan Chen
Martin Minařík, Melinda S. Modrell, J. Andrew Gillis, Alexander S. Campbell, Isobel Fuller, Rachel Lyne, Gos Micklem, David Gela, Martin Pšenička, Clare V. H. Baker
Min Feng, Baizhen Gao, L. Rene Garcia, Qing Sun
Panagiotis Tsimpos, Simon Desiderio, Pauline Cabochette, Sadia Kricha, Eric J. Bellefroid
| Stem cells, regeneration & disease modelling
Diffusible fraction of niche BMP ligand safeguards stem-cell differentiation
Sharif M Ridwan, Samaneh Poursaeid, Emma Kristine Beard, Autumn Twillie, Muhammed Burak Bener, Matthew Antel, Ann E Cowan, Shinya Matsuda, Mayu Inaba
Patrick M Helbling, Anjali Vijaykumar, Alvaro Gomariz, Karolina A Zielinska, Thomas Zerkatje, Kathrin Loosli, Stephan Isringhausen, Takashi Nagasawa, Ingo Roeder, Markus G Manz, Tomomasa Yokomizo, Cesar Nombela-Arrieta
Jimena Montagne, Matías Preza, Uriel Koziol
HIF1A contributes to the survival of aneuploid and mosaic pre-implantation embryos
Estefania Sanchez-Vasquez, Marianne E. Bronner, Magdalena Zernicka-Goetz
Léa Torcq, Sara Majello, Catherine Vivier, Anne A. Schmidt
James L. Engel, Xianglong Zhang, Daniel R. Lu, Olaia F. Vila, Vanessa Arias, Jasper Lee, Christopher Hale, Yi-Hsiang Hsu, Chi-Ming Li, Roland S. Wu, Vasanth Vedantham, Yen-Sin Ang
Sosuke Fujita, Mako Takahashi, Gaku Kumano, Erina Kuranaga, Masayuki Miura, Yu-ichiro Nakajima
Cellular senescence promotes progenitor cell expansion during axolotl limb regeneration
Qinghao Yu, Hannah E. Walters, Giovanni Pasquini, Sumeet Pal Singh, Martina Lachnit, Catarina Oliveira, Daniel León-Periñán, Andreas Petzold, Preethi Kesavan, Cristina Subiran, Ines Garteizgogeascoa, Dunja Knapp, Anne Wagner, Andrea Bernardos, María Alfonso, Gayathri Nadar, Alwin M. Graf, Konstantin E. Troyanovskiy, Andreas Dahl, Volker Busskamp, Ramón Martínez-Máñez, Maximina H. Yun
Yoko Nakai-Futatsugi, Jianshi Jin, Taisaku Ogawa, Noriko Sakai, Akiko Maeda, Ken-ichi Hironaka, Masakazu Fukuda, Hiroki Danno, Yuji Tanaka, Seiji Hori, Katsuyuki Shiroguchi, Masayo Takahashi
Folate depletion alters mouse trophoblast stem cell regulation in vitro
Joanna Rakoczy, Erica D Watson
Laura Celotto, Fabian Rost, Anja Machate, Juliane Bläsche, Andreas Dahl, Anke Weber, Stefan Hans, Michael Brand
Simon Perrin, Cécile-Aurore Wotawa, Vincent Bretegnier, Marine Luka, Fanny Coulpier, Cécile Masson, Mickael Ménager, Céline Colnot
Cátia Carvalho, Daniel J. Barbosa, Ricardo Celestino, Esther Zanin, Ana Xavier Carvalho, Reto Gassmann
Seungmae Seo, Sagar L. Patil, Yong-Oon Ahn, Jacqueline Armetta, Everardo Hegewisch-Solloa, Micah Castillo, Nicole C. Guilz, Achchhe Patel, Barbara Corneo, Malgorzata Borowiak, Preethi Gunaratne, Emily M. Mace
Evan W. Craig, Erik C. Black, Camille E.A. Goo, Avery Angell Swearer, Nathaniel G. Yee, Jeffrey P. Rasmussen
The Chordate Origins of Heart Regeneration
Keaton J. Schuster, Lionel Christiaen
Biallelic variants in LARS1 induce steatosis in developing zebrafish liver via enhanced autophagy
Masanori Inoue, Wulan Apridita Sebastian, Shota Sonoda, Hiroaki Miyahara, Nobuyuki Shimizu, Hiroshi Shiraishi, Miwako Maeda, Kumiko Yanagi, Tadashi Kaname, Reiko Hanada, Toshikatsu Hanada, Kenji Ihara
Yan Xue, Yiming Chao, Xinyi Lin, Yuanhua Huang, Joshua WK Ho, Ryohichi Sugimura
HLA-Based Banking of Human Induced Pluripotent Stem Cells in Saudi Arabia
Maryam Alowaysi, Robert Lehmann, Mohammad Al-Shehri, Moayad Baadheim, Hajar Alzahrani, Doaa Aboalola, Asima Zia, Dalal Malibari, Mustafa Daghestani, Khaled Alghamdi, Ali Haneef, Dunia Jawdat, Fahad Hakami, David Gomez-Cabrero, Jesper Tegner, Khaled Alsayegh
| Plant development
Transcriptional modulation during photomorphogenesis in rice seedlings
Parul Gupta, Pankaj Jaiswal
Carotenoid metabolism negatively regulates auxin-mediated root growth
Kang Xu, Haoran Zeng, Emi Yumoto, Masashi Asahina, Ken-ichiro Hayashi, Hidehiro Fukaki, Hisashi Ito, Masaaki K. Watahiki
Ethan J Redmond, James Ronald, Seth J Davis, Daphne Ezer
Haonan Bao, Rui Sun, Megumi Iwano, Yoshihiro Yoshitake, Shiori S Aki, Masaaki Umeda, Ryuichi Nishihama, Shohei Yamaoka, Takayuki Kohchi
Cellular gibberellin dynamics govern indeterminate nodule development, morphology and function
Colleen Drapek, Nadiatul A. Radzman-Mohd, Annalisa Rizza, Katharina Schiessl, Fabio Dos Santos Barbosa, Jiangqi Wen, Giles E.D. Oldroyd, Alexander M. Jones
A Single-Nucleus Atlas of Seed-to-Seed Development in Arabidopsis
Travis A. Lee, Tatsuya Nobori, Natanella Illouz-Eliaz, Jiaying Xu, Bruce Jow, Joseph R. Nery, Joseph R. Ecker
Investigating the genetic control of plant development under speed breeding conditions
Nicola Rossi, Wayne Powell, Ian Mackay, Lee Hickey, Andreas Maurer, Klaus Pillen, Karen Halliday, Rajiv Sharma
Edwin R Lampugnani, Staffan Persson, Ghazanfar Abbas Khan

Arabidopsis CML13 and CML14 Have Essential And Overlapping Roles In Plant Development
Kyle Symonds, Howard Teresinski, Bryan Hau, David Chiasson, Wayne A. Snedden
Nathaly Maldonado-Taipe, Elodie Rey, Mark Tester, Christian Jung, Nazgol Emrani
Travis Parker, Tayah Bolt, Troy Williams, Ramachandra Varma Penmetsa, Mwiinga Mulube, Antonia Palkovic, Celestina Nhagupana Jochua, Maria del Mar Rubio Wilhelmi, Sassoum Lo, Gail Bornhorst, Li Tian, Kelvin Kamfwa, Sam Hokin, Andrew Farmer, Christine H. Diepenbrock, Paul Gepts
Differential mutation accumulation in plant meristematic layers
Kirk R Amundson, Mohan Prem Anand Marimuthu, Oanh Nguyen, Konsam Sarika, Isabelle J DeMarco, Angelina Phan, Isabelle M Henry, Luca Comai
From buds to shoots: Insights into grapevine development from the Witch’s Broom bud sport
Eleanore J. Ritter, Peter Cousins, Michelle Quigley, Aidan Kile, Sunil K. Kenchanmane Raju, Daniel H. Chitwood, Chad Niederhuth
Cristina M Alexandre, Kerry L Bubb, Karla M Schultz, Janne Lempe, Josh T Cuperus, Christine Queitsch
João R. D. Ramos, Blanca Jazmin Reyes-Hernández, Karen Alim, Alexis Maizel
Viviana June, Xiaoya Song, Z. Jeffrey Chen
Phosphate starvation regulates cellulose synthesis to modify root growth
Ghazanfar Abbas Khan, Arka Dutta, Allison Van de Meene, Kristian EH. Frandsen, Michael Ogden, James Whelan, Staffan Persson
DEVIL peptides control cell growth and differentiation in different developmental processes
Ana Alarcia, Amparo Primo-Capella, Elena Perpiñán, Priscilla Rossetto, Cristina Ferrándiz

Ansar Ali, Chi Kuan, Fu-Yu Hung, Tsai-Chen Chen, Hui-Chun Lee, Shao-Li Yang, Yun-Ru Feng, Keqiang Wu, Chin-Min Kimmy Ho
Expression of cell-wall related genes is highly variable and correlates with sepal morphology
Diego A. Hartasánchez, Annamaria Kiss, Virginie Battu, Charline Soraru, Abigail Delgado-Vaquera, Florian Massinon, Marina Brasó-Vives, Corentin Mollier, Marie-Laure Martin-Magniette, Arezki Boudaoud, Françoise Monéger
| Evo-devo
Control of cell fate specification and patterning by an ancestral microRNA
Adolfo Aguilar-Cruz, Eduardo Flores-Sandoval, Ximena Gutiérrez-Ramos, Omar Oltehua- Lopez, Ana E. Dorantes-Acosta, Joshua T. Trujillo, Hirotaka Kato, Kimitsune Ishizaki, Rebecca A. Mosher, Liam Dolan, Daniel Grimanelli, Jim Haseloff, John L. Bowman, Mario A. Arteaga-Vazquez
Zhiliang Zhang, Zhifei Zhang, Lars E. Holmer, Timothy P. Topper, Bing Pan, Guoxiang Li
Insights into Digit Evolution from a Fate Map Study of the Forearm
JDH Oh, DDZ Saunders, L McTeir, M Jackson, JD Glover, JJ Schoenebeck, LA Lettice, MG Davey
Emily E. K. Kopania, Gregg W. C. Thomas, Carl R. Hutter, Sebastian M. E. Mortimer, Colin M. Callahan, Emily Roycroft, Anang S. Achmadi, William G. Breed, Nathan L. Clark, Jacob A. Esselstyn, Kevin C. Rowe, Jeffrey M. Good
E Lafuente, D Duneau, P Beldade
Uncovering the mosaic evolution of carnivoran skeletal systems
Chris J. Law, Leslea J. Hlusko, Z. Jack Tseng
J.P. Lyons, K.D. Kavanagh
Xavier Grau-Bové, Lucie Subirana, Lydvina Meister, Anaël Soubigou, Ana Neto, Anamaria Elek, Oscar Fornas, Jose Luis Gomez-Skarmeta, Juan J Tena, Manuel Irimia, Stéphanie Bertrand, Arnau Sebé-Pedrós, Hector Escriva
Adaptive Functions of Structural Variants in Human Brain Development
Wanqiu Ding, Xiangshang Li, Jie Zhang, Mingjun Ji, Mengling Zhang, Xiaoming Zhong, Yong Cao, Xiaoge Liu, Chunqiong Li, Chunfu Xiao, Jiaxin Wang, Ting Li, Qing Yu, Fan Mo, Boya Zhang, Jianhuan Qi, Jie-Chun Yang, Juntian Qi, Lu Tian, Xinwei Xu, Qi Peng, Wei-Zhen Zhou, Zhijin Liu, Aisi Fu, Xiuqin Zhang, Jian-Jun Zhang, Yujie Sun, Baoyang Hu, Ni A. An, Li Zhang, Chuan-Yun Li
Hybridization and its impact on the ontogenetic allometry of skulls in macaques
Tsuyoshi Ito, Ryosuke Kimura, Hikaru Wakamori, Mikiko Tanaka, Ayumi Tezuka, Atsushi J. Nagano, Yuzuru Hamada, Yoshi Kawamoto
Yuting Liu, Xin Luo, Yiming Sun, Kaimin Chen, Ting Hu, Benhui You, Jiahao Xu, Fengyun Zhang, Xiaoyu Meng, Xiang Li, Xiechao He, Cheng Li, Bing Su
Gene regulatory network that shaped the evolution of larval sensory organ in Cnidaria
Eleanor Gilbert, Jamie Craggs, Vengamanaidu Modepalli
Juan Jose Lagos-Oviedo, Ido Pen, Jan Jakob Kreider
Cell Biology
Cerebellar granule cell migration and folia development requires Mllt11/Af1q
Marley Blommers, Danielle Stanton-Turcotte, Emily A. Witt, Mohsen Heidari, Angelo Iulianella
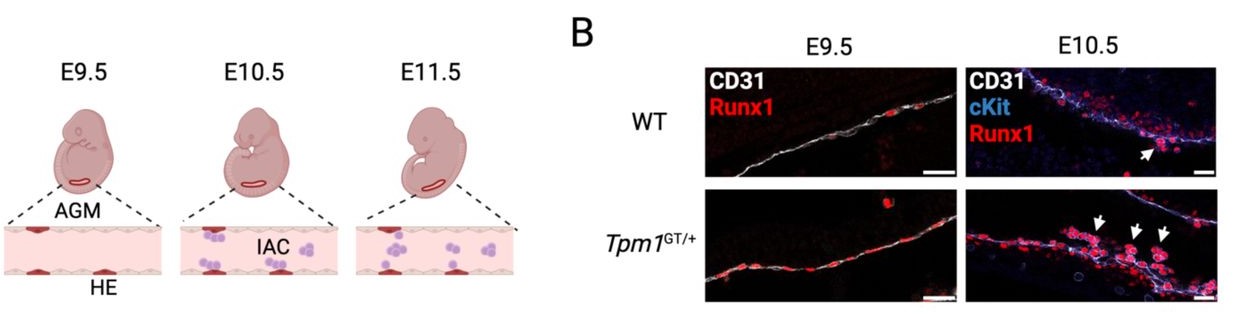
Madison B Wilken, Gennadiy Fonar, Catriana Nations, Giulia Pavani, Victor Tsao, James Garifallou, Joanna Tober, Laura Bennett, Jean Ann Maguire, Alyssa Gagne, Nkemdilim Okoli, Paul Gadue, Stella T Chou, Nancy A Speck, Deborah L French, Christopher S Thom
Kelsey R. Clearman, Napassawon Timpratoom, Dharti Patel, Addison B. Rains, Courtney J. Haycraft, Mandy J. Croyle, Jeremy F. Reiter, Bradley K. Yoder
Yongchun Zhang, Dimitris Karagiannis, Helu Liu, Mi Lin, Yinshan Fang, Ming Jiang, Xiao Chen, Supriya Suresh, Haidi Huang, Junjun She, Feiyu Shi, Patrick Yang, Wael El-Rifai, Alexander Zaika, Anthony E. Oro, Anil K. Rustgi, Timothy C. Wang, Chao Lu, Jianwen Que
Soraya Villaseca, Juan Ignacio Leal, Jossef Guajardo, Hernan Morales-Navarrete, Roberto Mayor, Marcela Torrejón
Naoki Kubo, Ryuji Uehara, Shuhei Uemura, Hiroaki Ohishi, Kenjiro Shirane, Hiroyuki Sasaki
Moawiah M Naffaa, Henry H. Yin
Edgar M. Pera, Josefine Nilsson-De Moura, Yuriy Pomeshchik, Laurent Roybon, Ivana Milas
An architectural role of oskar mRNA in granule assembly
Mainak Bose, Branislava Rankovic, Julia Mahamid, Anne Ephrussi
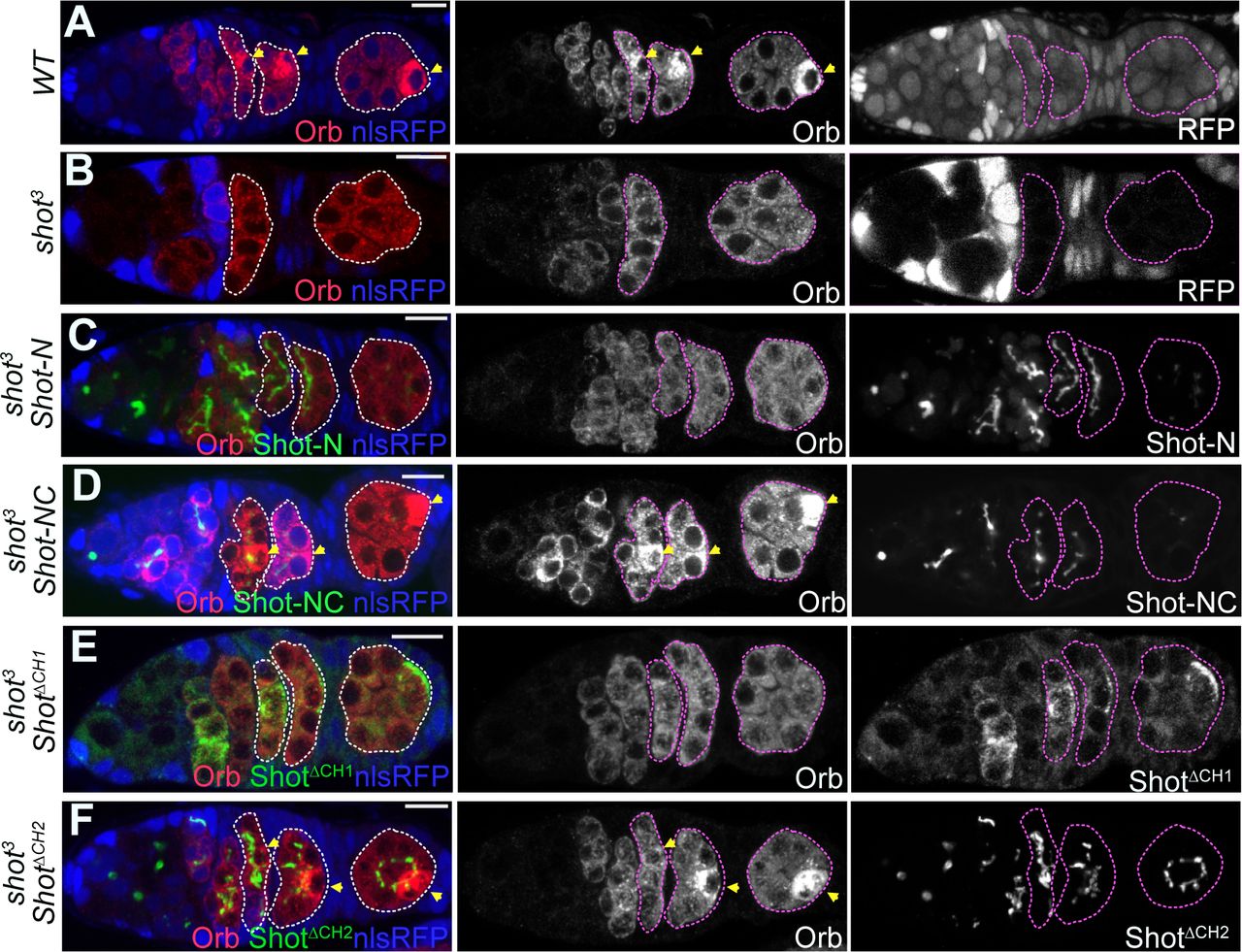
The Shot CH1 domain recognises a distinct form of F-actin during Drosophila oocyte determination
D. Nashchekin, I. Squires, A. Prokop, D. St Johnston
Modelling
Morphogen gradients can convey position and time in growing tissues
Roman Vetter, Dagmar Iber
A dynamic Hedgehog gradient orients tracheal cartilage rings
Evan P. Kingsley, Darcy Mishkind, Tom W. Hiscock, Clifford J. Tabin
Plasticity-led evolution as an intrinsic property of developmental gene regulatory networks
Eden Tian Hwa Ng, Akira R. Kinjo
Mechanistic Regulation of Planarian Shape During Growth and Degrowth
Jason M. Ko, Waverly Reginato, Daniel Lobo
Tools & Resources
A Meta-Atlas of the Developing Human Cortex Identifies Modules Driving Cell Subtype Specification
Patricia R Nano, Elisa Fazzari, Daria Azizad, Claudia V Nguyen, Sean Wang, Ryan L Kan, Brittney Wick, Maximilian Haeussler, Aparna Bhaduri
Fides Zenk, Jonas Simon Fleck, Sophie Martina Johanna Jansen, Bijan Kashanian, Beneditk Eisinger, Malgorzata Santel, Jean Samuel Dupre, Gray Camp, Barbara Treutlein
Mapping the developmental potential of mouse inner ear organoids at single-cell resolution
Joerg Waldhaus, Linghua Jiang, Liqian Liu, Jie Liu, Robert Keith Duncan
Germ cells do not progress through spermatogenesis in the infertile zebrafish testis
Andrea L. Sposato, Darren R. Llewellyn, Jenna M. Weber, Hailey L. Hollins, Madison N. Schrock, Jeffrey A. Farrell, James A. Gagnon
A single-cell atlas of pig gastrulation as a resource for comparative embryology
Luke Simpson, Andrew Strange, Doris Klisch, Sophie Kraunsoe, Takuya Azami, Daniel Goszczynski, Triet Le, Benjamin Planells, Nadine Holmes, Fei Sang, Sonal Henson, Matthew Loose, Jennifer Nichols, Ramiro Alberio
Single-cell transcription mapping of murine and human mammary organoids responses to female hormones
Jenelyz Ruiz-Ortiz, Steven M. Lewis, Michael F. Ciccone, Deeptiman Chatterjee, Samantha Henry, Adam Siepel, Camila O. Dos Santos
Emily Burghardt, Jessica Rakijas, Antariksh Tyagi, Pralay Majumder, Bradley J.S.C. Olson, Jocelyn A. McDonald
A Window into Mammalian Basement Membrane Development: Insights from the mTurq2-Col4a1 Mouse Model
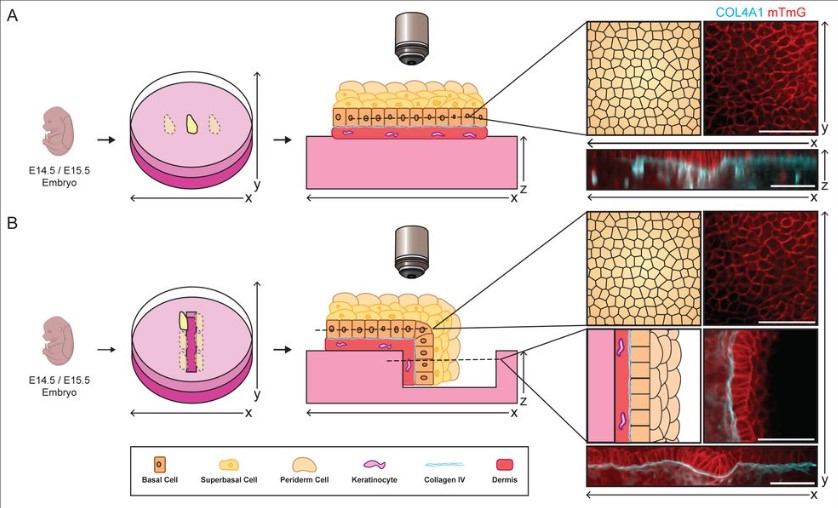
Rebecca A. Jones, Brandon Trejo, Parijat Sil, Katherine A. Little, H. Amalia Pasolli, Bradley Joyce, Eszter Posfai, Danelle Devenport
Platre Matthieu Pierre, Mehta Preyanka, Halvorson Zachary, Ling Zhang, Brent Lukas, Gleason F. Matias, Faizi Kian, Goulding Callum, Busch Wolfgang
Research practice & education
Carmen Herrera Sandoval, Christopher Borchers, Scott Takeo Aoki
Andrea M. Henle
Controlled experiment finds no detectable citation bump from Twitter promotion
Trevor A. Branch, Isabelle M. Cȏté, Solomon R. David, Joshua A. Drew, Michelle LaRue, Melissa C. Márquez, E. Chris M. Parsons, D. Rabaiotti, David Shiffman, David A. Steen, Alexander L. Wild
Kelly D. Cobey, Sanam Ebrahimzadeh, Matthew J. Page, Robert T. Thibault, Phi-Yen Nguyen, Farah Abu-Dalfa, David Moher


 (No Ratings Yet)
(No Ratings Yet)

 (5 votes)
(5 votes)