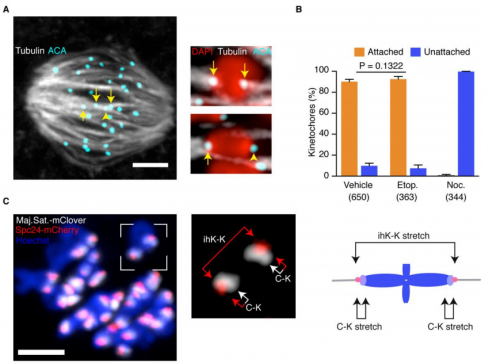
This interview by Aidan Maartens appeared in Development, Vol 144 Issue 19
Jayaraj (Jay) Rajagopal is a Principal Investigator at the Center for Regenerative Medicine at Massachusetts General Hospital and an Associate Professor of Medicine at Harvard Medical School. A Howard Hughes Medical Institute Faculty Scholar, his lab works on the development and regeneration of the lung. He uses stem cell and animal models to develop novel insights that hopefully will provide inspiration for therapies to help treat human lung disease. He was awarded the Dr Susan Lim Award for Outstanding Young Investigator at the 2017 International Society for Stem Cell Research (ISSCR) meeting in Boston (MA,USA), where we met him to talk about how a fish tank started a life-long fascination with the lung, the transition to running his own lab, and his optimism for the future of both basic stem cell research and its clinical translation.

You’re here in Boston to collect ISSCR’s Susan Lim Young Investigator award: what does the award mean to you?
It really was an incredible honour. I looked over the past recipients and the vast majority of them are either friends or people whose work I admire. Additionally, Susan Lim and her husband Deepak Sharma are just wonderful people – it was great to meet them, and especially their daughter who is interested in becoming a physician. Most of all, it represents a chance to be embedded in a rich community of scholarship that promotes education and younger researchers.
What got you interested in science in the first place?
That started very early for me: I was always intrigued by animals. I spent my summers in India as a child and when all my cousins were in school I’d be out on the farm, capturing animals and things like that. The other early influence was my father, who was a doctor and would talk with me about interesting cases. His perspective was great because he introduced these diseases as fascinating mysteries. For as long as I can remember, those two things – animals and medicine – were what I wanted to devote my professional life to.
How did you become interested in developmental biology and the development of the lung in particular?
I went to college at Harvard and studied molecular biology, and actually specifically avoided developmental biology! Having read The Eighth Day of Creation by Horace Freeland Judson, molecular biology seemed so exciting. I worked with Jack Szostak and Jenn Doudna at the time when self-splicing RNAs were new. We could synthesise the RNA molecules in vitro, put them in a solution and test their enzymatic activity with exquisite precision. The whole process was a lot of fun and appealingly very understandable. Then I went to medical school and loved the human biology and physiology, as well as being able to interact with and treat patients. But at some point I stepped back and asked myself: what do I love? I realised that, at heart, I like cells and tissues, and how they come together functionally to create a living animal, essentially exactly what I was fascinated by as a kid in a more mature guise. Developmental biology just seemed like the natural thing to do.
My interest in the lung also traces back to when I was young. I had a fish tank and even as a very young kid I was mesmerised by it: these animals could breathe underwater, but if you let your tank get dirty – like many kids do – many fish can also breathe air. In medical school I learned about the lung’s physiology, and how beautifully it works to bring oxygen to the bloodstream. When I was looking for a lab to join in Boston, there wasn’t anyone studying the lung in the way that I wanted to. Doug Melton was just getting interested in the pancreas, moving from his more basic work in the frog, and I thought that since the lung and the pancreas came from the same tube, I could help him figure out how to make a pancreas, and then once I was done with my training in his lab, I’d move a little anterior in the tube and make a lung. Doug thought it was a great idea, and we hit it off immediately. The best laid plans often change though. When induced pluripotent stem cells (iPSCs) came out, I immediately started working on making iPSCs into β-cells, and that got me hooked on stem cells. Doug really was a fantastic mentor to me, broadening my perspectives, encouraging me to explore the newest ideas and systems, while maintaining my fundamental interests.
What were your aims when you first started your lab?
As part of my interview at the Center for Regenerative Medicine at Massachusetts General Hospital I gave a chalk talk describing my ideas, but my actual research program ended up only very loosely related to that chalk talk. Essentially, I didn’t really know what I was going to do, except in very vague terms, and although I was empowered with new experimental tools and an understanding of the basis of experimentation, I was left with pretty much the same interests I had as a child: how do organs come to be?
When my lab started I was initially not so interested in the iPSC differentiation paradigms I had explored in Doug’s lab. Rather, I was enamoured of the developmental biology of the lung and particularly of tissue regeneration – it was so beautiful, and that’s what I focused on, primarily in the mouse because the biological tools were much more rigorous. Yet at the same time, I had always had this keen interest in human basic biology and in medicine – so I sort of put myself in Doug’s shoes again and eventually realised I would have to return to iPSCs. We reinvigorated that aspect of the lab, and then moved on to developing systems to grow adult lung stem cells, and I think we now have a way to use an actual human lung explant to do what I’ve always wanted to do: investigate the developmental biology of a regenerating human mini-organ.
What is it about the lung that is so fascinating as a developmental system?
For me one reason is evolution, which was another interest of mine since I was a kid. Back to my fishtank: it’s a mystery how fish came out of the water. How did the first lungs arise? The question of how it came about is fascinating. It’s also a fascinating organ in terms of its physiology, which I was introduced to during my medical studies. And something really appealing about the lung is that it is an organ par excellence for understanding regeneration. You have to breathe – which means there are physical forces and gas fluxes, and you inhale allergens, infections, toxins, dust and so on. All of the cardinal ways in which the environment could possibly perturb a tissue are captured in the lung. We’re really interested in how a perturbation in a tissue wrought by one of these injuries is resolved. I do often wonder whether there are some forms of simple equations that you could write to understand how cells come together to generate ensemble properties of tissues. It’s a very tall order, but there has got to be at least partially ordered ways of thinking about it, some ‘laws’ of regeneration.
And the lung is turning out to be a lot more complex than we previously thought. Doing single cell sequencing in collaboration with Aviv Regev we find two things: firstly, that there’s a whole new set of cells with interesting physiological properties; and secondly that many cells have specific immune signatures. The epithelium was considered to be pretty monotonous, and to interact with a panoply of weird different immune cells (you can see I’m not an immunologist!). Now it looks like there may be considerable crosstalk of particular epithelial cells types to particular types of immune cells. We also learned there are many subtypes of known cells, totally novel cells important for disease, and even new specialised structures in the airway.
For me it seemed like everything came together in the lung: aesthetics, evolution, developmental biology, the clinical aspect, and how the organ interacts with the environment. Somehow you just find your groove if you keep doing what you like, and the lung is a perfect lens with which to ask all the questions I’ve ever wanted to ask. I understand the developmental biology and some of the therapeutic issues, and then we are collaborating with people from other disciplines who look at the problem through different lenses. Some are computational biologists, physicists with new imaging techniques, immunologists, and I’m interested in collaborating with a whole suite of other biologists, including neuroscientists, evolutionary biologists and mathematical modellers. I don’t think that any single type of biologist is going to get close to sorting out the ‘laws’ of tissue behaviour unless they work together. It’s one of the most fun things in science: to head into totally new domains. It’s also a good way to make new friends! I get bored relatively easily and so like constantly hopping from one aspect of tissue biology to another: from evolution to signalling to force transduction to hypoxia. And I suspect many of these topics will become reincarnated in the lab as a new idea makes one of them seem appealing again. My tendency to dalliance works well for the postdocs as they can take their own projects with them to their own labs.
The lung is a perfect lens with which to ask all the questions I’ve ever wanted to ask
Many speakers at this meeting have emphasised the importance of developmental biology for stem cell research – how do you see the relationship between the two fields?
One way stem cell biology has helped developmental biology is that developmental biologists were so interested in embryogenesis, but tended not to think about the adult organism. Stem cell biology made developmental biologists think about the adult. The idea of stem cells is also just useful to convey enthusiasm, and that enthusiasm is incredibly important. Even in my own case, stem cells drew me into the problem of lung development and made me think about it differently. Stem cells are super interesting – but they are only one aspect of tissue function. There was a quote from Jean Rostand on the wall of Doug Melton’s lab which said ‘Theories come and go, but the frog remains’. I think this serves to remind us that there are tissues, organs and animals. Those exist, and all these different ways of doing science and naming cell functions are just different lenses with which to look at them, none of them complete in their own right and always evolving. As we analyse cells more deeply in the lung epithelium, we are finding that none is associated with a unitary specific function, but they all have many distinct activities. Stem cells aren’t just for replication and differentiation anymore. We have shown that they signal and I am sure they sense and perform necessary metabolic activities too.
It’s also important not to let the translational side eclipse everything else. The constant translational need, which is important, can become a drumbeat that moves you away from basic biology. Stem cells have this immediate connotation of being of use in a practical fashion, which is part of what makes them wonderful, but developmental biology has always come from the first principles of ‘I would just like to understand something’, and I think we cannot lose that because most of our truly game-changing ideas are a result of curiosity-driven basic biology.
Finally, stem cell research is an easy paradigm to communicate to patients and their families. It’s much harder to educate society at large about the interest and importance of basic biology. But I think we have to do that, especially with the modern political climate – there can’t be anything more important than to just explain to people why new knowledge is important.
So do you think the stem cell field as a whole is good at engaging with the public?
First of all, I think what the ISSCR does is spectacular, and I would in particular point out people like George Daley, Len Zon, David Scadden and Doug Melton, who are all great communicators, not just to the public at large, but also to our politicians. From my own perspective and background, I think we need to think about educating patients themselves, and to remember that they are vulnerable. I was a pulmonologist, and if I had a patient with, let’s say, idiopathic pulmonary fibrosis, they would walk into the office short of breath. More or less the one thing I could do for them was to give them oxygen – that is where the treatment ended. So you can imagine my inbox was full of messages from people who couldn’t get a new functioning lung with a lung transplant. These patients all wanted ‘stem cells’. I’m usually very straightforward in my response to them: although I empathise with what they have to go through because I’ve seen it first hand, I tell them that currently, there are no stem cell therapies for the lung, and that I would be very careful because there are a lot of people squirting stem cells into people without the proper science. But at the same time I try to give them the optimism of research, because I really do believe in it, and I am only getting more optimistic about it these days. I really think there is going to be a renaissance in terms of therapies produced through iPSCs and organoid models, and I try to fill patients with that kind of optimism. Unfortunately, you can’t give patients a timetable when they are desperate for a cure, but I can honestly say the research keeps moving faster and faster than I could ever have imagined.
At one point in my career, we wanted to make iPSCs from cystic fibrosis patients so that we could model the disease. I talked to some of my clinician friends, and they agreed to ask some of their patients to contribute, and the result was unbelievable – within a couple of weeks, we were turning people away. It’s overwhelming to see that kind of enthusiasm from patients: even if you tell them that this donation is very unlikely to help them personally, they still want to contribute. It’s also reciprocal: one of my graduate students working on a cystic fibrosis-related project asked me if he could meet a patient. My clinical friends again had a patient lined up to meet him before you could imagine it, and that patient spoke to my laboratory. It just blew them away and made their research so much more meaningful. If you are a PhD scientist working on a disease at the bench, you’re missing something. If you’ve come to meet patients, when an experiment fails 90 times, there is another reason you’re repeating it – it’s not just that you want the answer, but there’s a person that needs your help.
Being a PI was just intrinsically so much more fun for me than being a postdoc
When you started your lab, what was the transition to being a PI like?
I have to say that I found my post-doctoral fellowship very hard. I came from clinical medicine, and was used to knowing what to do – even if you couldn’t save every patient, you knew you could do your job well, and in a well-defined way. In science, it didn’t work that way and, quite frankly, it took me a long time to learn how to fail, and to appreciate the importance of problem solving. I am actually one of those scientists that did not love bench work, but rather I preferred to look at and think about data, talk to colleagues about new ideas and dream up future experiments. I had also always known, since I had been a Chief Resident in Internal Medicine at Massachusetts General Hospital, that mentoring was my absolute favourite thing. So it turned out that being a PI was just intrinsically so much more fun for me than being a postdoc, for almost every single reason. Again, I’d found my groove – being a laboratory head is the job I was ‘supposed’ to do, and it has only gotten better and better and better. I can’t imagine a job that is more fun, and I think the core value I treasure about it, perhaps even more than the study of living things, is the freedom it provides me.
One great thing about lung science for me is that, unlike haematopoiesis, neuroscience or immunology, it is so understudied, and so it’s just a great place for me to train young scientists. I’m so pleased to say that the first four scientists to have graduated from my lab all have their own labs. I don’t think I’ll ever have a very big lab – I like to put a lot of energy into every single postdoc and student, and hopefully have all of them leave and still love biology, no matter what they decide to do. That said, I’m hoping every single one will have their own laboratory if that is what appeals to them. Also, thanks to the Harvard Stem Cell Institute and my relationship to the undergraduate Stem Cell and Regenerative Biology Department at Harvard University, we always have undergraduates in the lab. It’s just fantastic to have people at every single stage of their education and career. Having undergraduates in my lab also means my postdocs and graduate students have a mentee, which is also a crucial part of growing as a scientist.
Do you have any advice for someone thinking about a scientific career today?
There have been editorials written by very prominent scientists that have said that we are training too many PhDs; I completely disagree with this! If anything I would say we are training too few, because while you can look at a PhD as a means towards one particular end, you could also look at it as a pure form of education that trains you to think and problem solve, and also to communicate in various ways. In some sense it’s a liberal arts education in terms of problem solving. But if you gravitated towards writing you could become a wonderful editor, if you gravitated towards ethics, politics or law, you can go into those roles. You can go into clinical medicine: a doctor who understands science is very valuable, just like a scientist who understands medicine. I remember in my own time, it used to be considered wrong to go to a company; now there are brilliant scientists who want to go to a company from day one. I love being a PI, and it’s a path I want to encourage but I’d like to see my young people in diverse spheres of human activity that captivate their imagination. The more people we have doing different things, but bound by a respect for scientific inquiry, the better.
I would say to a young person: explore and try to figure out what you love, and don’t worry too much about exactly where you’ll be 20 years from now because, in my view, no one really knows. Life is an experiment with no controls – at some level we’ll never know whether we’ve taken the right path – but if you’re constantly doing something you’re excited about, work becomes play. If you can live a life where work is always play, you have sealed the deal professionally. On top of that, you have to think about work-life balance – different paths have different constraints, and it’s important to talk to people who have lives that you’d want to emulate at work and also at home. Similarly with money: different people have different thresholds concerning what they are comfortable earning, and it’s important to think about this (but money can’t buy you love). I’d also advise people to ask for advice and help from people they respect. Mentors who listen and empathise, and who have a solid sense for your abilities can be invaluable for you as they were for me. I’ve found many people believed in my ultimate success and that was enormously encouraging at times when I did not think I could rise to the level of my own scientific bar.
Is there anything that Development readers would be surprised to find out about you?
Well the things I love outside of the lab are music, reading, the arts, travel and my family. I guess they’re all about exploration, and that might be the common theme. It’s a pretty interesting life to be able to combine all of these things.
Perhaps the other thing is that I’ve never left Boston since coming here for college, where in fact I met my wife in my freshman year! I’ve always found Boston to be supportive – people often think of it as an intense and competitive environment, but I’ve found it to be completely the opposite. Whenever I’ve asked for help, I’ve gotten it!
 (1 votes)
(1 votes)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) (18 votes)
(18 votes) The Society for Developmental Biology is deeply concerned about the damage caused by Hurricanes Irma and Maria to the laboratories of our colleagues in Puerto Rico. In order to facilitate the continuation of research programs in those labs or at another temporary host location, SDB is offering relief grants of up to $10,000 to each lab. Funds may be used for replacement of organisms, reagents, supplies, travel to host lab by PI or his/her trainees, core facility usage fees at host institution, etc. For questions, please contact Ida Chow at
The Society for Developmental Biology is deeply concerned about the damage caused by Hurricanes Irma and Maria to the laboratories of our colleagues in Puerto Rico. In order to facilitate the continuation of research programs in those labs or at another temporary host location, SDB is offering relief grants of up to $10,000 to each lab. Funds may be used for replacement of organisms, reagents, supplies, travel to host lab by PI or his/her trainees, core facility usage fees at host institution, etc. For questions, please contact Ida Chow at  (3 votes)
(3 votes)