This article was first published in Development, and was written by Ben-Zion Shilo, Weizmann Institute of Science, Israel.
How can the revolution in our understanding of embryonic development and stem cells be conveyed to the general public? Here, I present a photographic approach to highlight scientific concepts of pattern formation using metaphors from daily life, displaying pairs of images of embryonic development and the corresponding human analogy. By making the viewer ‘feel’ like a cell within a developing embryo, the personal experiences resonate with the scientific concepts, facilitating a new type of appreciation.
The field of developmental biology has undergone a revolution in the past three decades. The conserved signaling pathways cells use to communicate with each other during embryogenesis have been elucidated. Recent technological developments include sophisticated genetic tools to interrogate the roles of genes in distinct tissues and specific time windows in a wide range of organisms, as well as new imaging technologies for the dynamic visualization of developmental processes at subcellular resolution. Finally, the identification of stem cells in different organs, and increasing understanding of the means by which these cells differentiate, brings hope that we might be able to recapitulate these processes for regenerative therapies.
This revolution affects us as scientists on a daily basis, but also has wider implications to every human being: from the understanding of how we are formed from embryos and how similar we are to other multicellular organisms, to the medical applications of this knowledge. When asking if the public is aware of these issues, I am disappointed to say that the answer to this question is no. This is mainly because existing channels to communicate these ideas are not sufficient, rather than reflecting a lack of interest in the topic. Any effort in broadening and enhancing public understanding of developmental biology is thus extremely important.
One challenge in presenting science in a popular way is the limited background knowledge of the audience. The presentation, be it a lecture, movie or article, tends to flow in only one direction: the expert ‘takes the audience by the hand’ and guides them through the scientific paradigms presented. By contrast, developing a dialog, whereby the audience would be able to resonate their own life experiences with the new scientific knowledge, might enhance the understanding of the principles and make the process more interactive and intuitive.
The cell is the basic unit in a developing organism, and it is the interaction between cells that drives the orderly process of embryonic development. In many respects, this concept can be compared to a human society, in which rules, communication and interactions between individuals lead to the intricate organization at physical and social levels. By basing a popular presentation on such an analogy, the audience immediately becomes actively involved. People can identify themselves with the developing cells and this association allows them to ‘feel’ the processes from a personal point of view. Interestingly, Robert Hooke first coined the term ‘cell’ in a biological context in 1665 as an analogy to the human world: the cell walls of an oak tree bark under his microscope reminded him of the cells of monks.
How can this analogy be elaborated and presented? In our research, we generate microscopic images that are not only highly informative to the researcher but also exceptionally beautiful. Yet, in the absence of sufficient background knowledge, these images can be largely meaningless to the general public. I have considered the prospects of utilizing such scientific images more effectively for public education, by using them as a tool for presenting analogies between the biological microscopic world and the macroscopic human one. Concentrating on common underlying principles between the two worlds, our intuitive understanding of the human world can be harnessed in order to grasp scientific paradigms of embryonic development.
During the academic year of 2011/2012, I was fortunate to be a fellow at the Radcliffe Institute for Advanced Study at Harvard University, and dedicated my time to developing this concept. The first step was to delineate the major principles of developmental biology in a logical and balanced manner. This process was extremely beneficial and involved many inspiring discussions with members of my lab and other colleagues. Then, with the major concepts in mind, I approached scientific colleagues working on different model organisms, and asked for images that would depict these concepts. It was rewarding to see how responsive the community was, and I ended up with dozens of images and movies, from which I chose the ones that displayed the concepts most clearly, and which I found most aesthetic.
As researchers, our eye is trained to focus on the crucial aspects of an elaborate scientific image, but for the naive observer such images are extremely confusing. For example, a bright auto-fluorescence spot that researchers ignore, may become their focus of attention. Michal Rovner, a renowned Israeli video artist, suggested converting the scientific images to black and white, leaving only the main topic in a single color. Indeed, it became immediately apparent that after this manipulation the scientific images become more effective: they are minimalistic, and appear more as a work of art. The subject ‘jumps out’ at the viewer, be it spaced neuroblasts (Fig. 1, top) or a stem cell niche (Fig. 2, top).
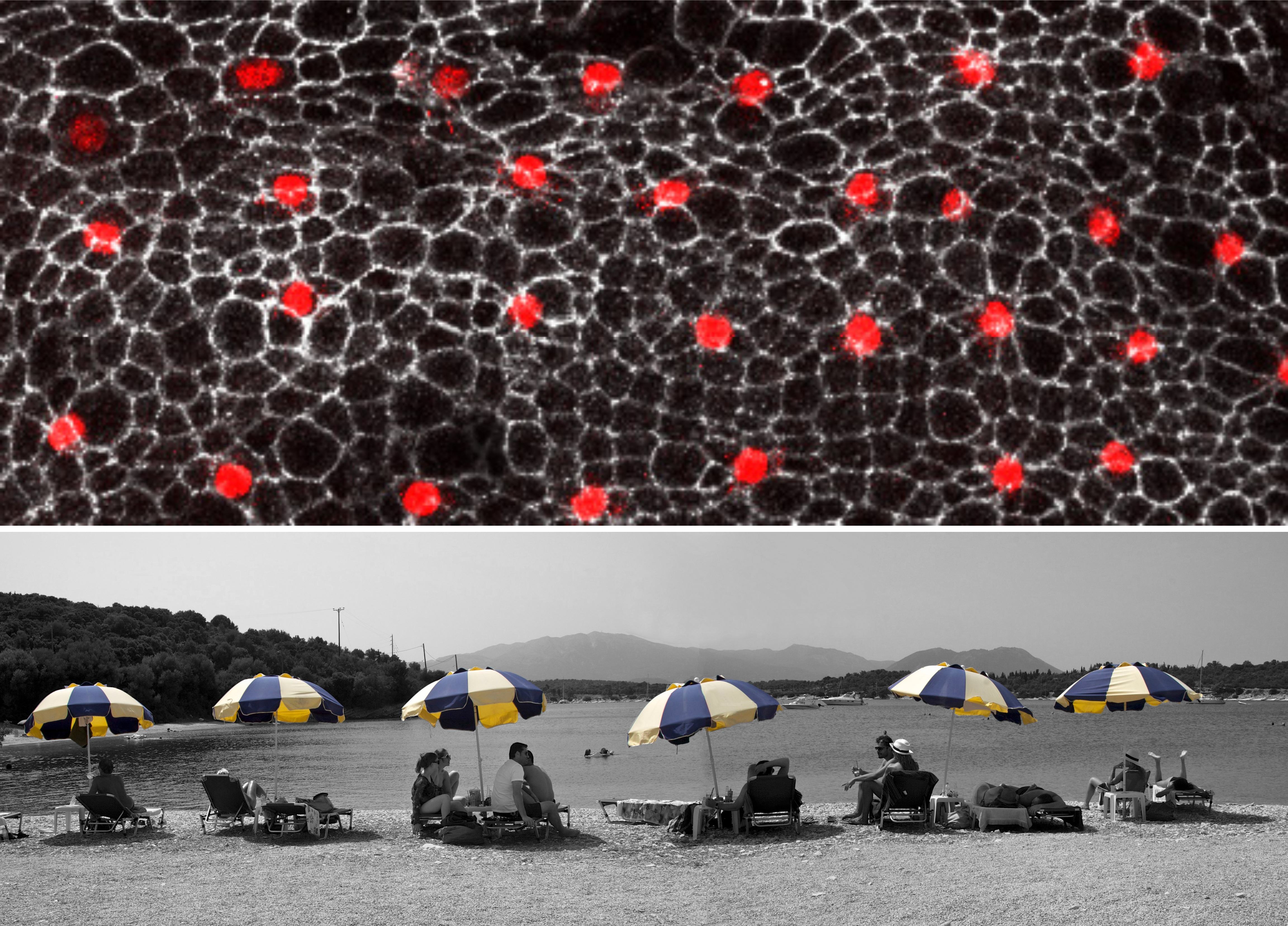
Figure 1- How cell signaling establishes a repeated pattern. The outer cell layer of the fruit fly pupa displays a regular arrangement of nerve (red) and epidermal (black) cells. All cells are initially equivalent to each other. Yet, random fluctuations in the level at which each cell produces a signal, allow a given cell to accumulate sufficiently high levels to make it a nerve cell. In parallel, this cell instructs its immediate neighbors to become skin cells. Eventually, an ordered array of nerve cells is generated. Top: external cell layer of the fruit fly pupa with regularly spaced nerve cells (red) (S. Yamamoto and H. Bellen, Baylor College of Medicine and Howard Hughes Medical Institute). Bottom: sunshades distributed at regular intervals on a beach at Lefkada island, Greece (B. Shilo). Note: the legends for Figs 1 and 2 represent the original explanations at the exhibit, and are aimed at the broad audience.
The next phase was to match the images with human metaphors. The process of thinking about the metaphors proved to be very creative and inspiring. Just like the procedure of defining the main scientific principles, finding the human metaphors was extremely revealing, forcing me to distill the most seminal aspects of scientific paradigms and choose the analogies to illustrate them best. I was mainly looking for a conceptual similarity rather than a visual one, but of course welcomed the possibility of having a visual resemblance. Thus, the regular spacing of sun umbrellas on the beach is both a visual and conceptual metaphor for the mechanisms underlying the patterning of neuroblasts in the Drosophila epidermis (Fig. 1), whereas the yeast ‘mother culture’ used for making bread has only a conceptual resonance to the idea of a stem cell niche (Fig. 2). Similarly, a flashing ‘Pizza’ sign becomes the metaphor for different times of exposure to the Sonic Hedgehog gradient in the limb, and a game of dominos resembles the successive elaboration of pattern in the embryonic axes.
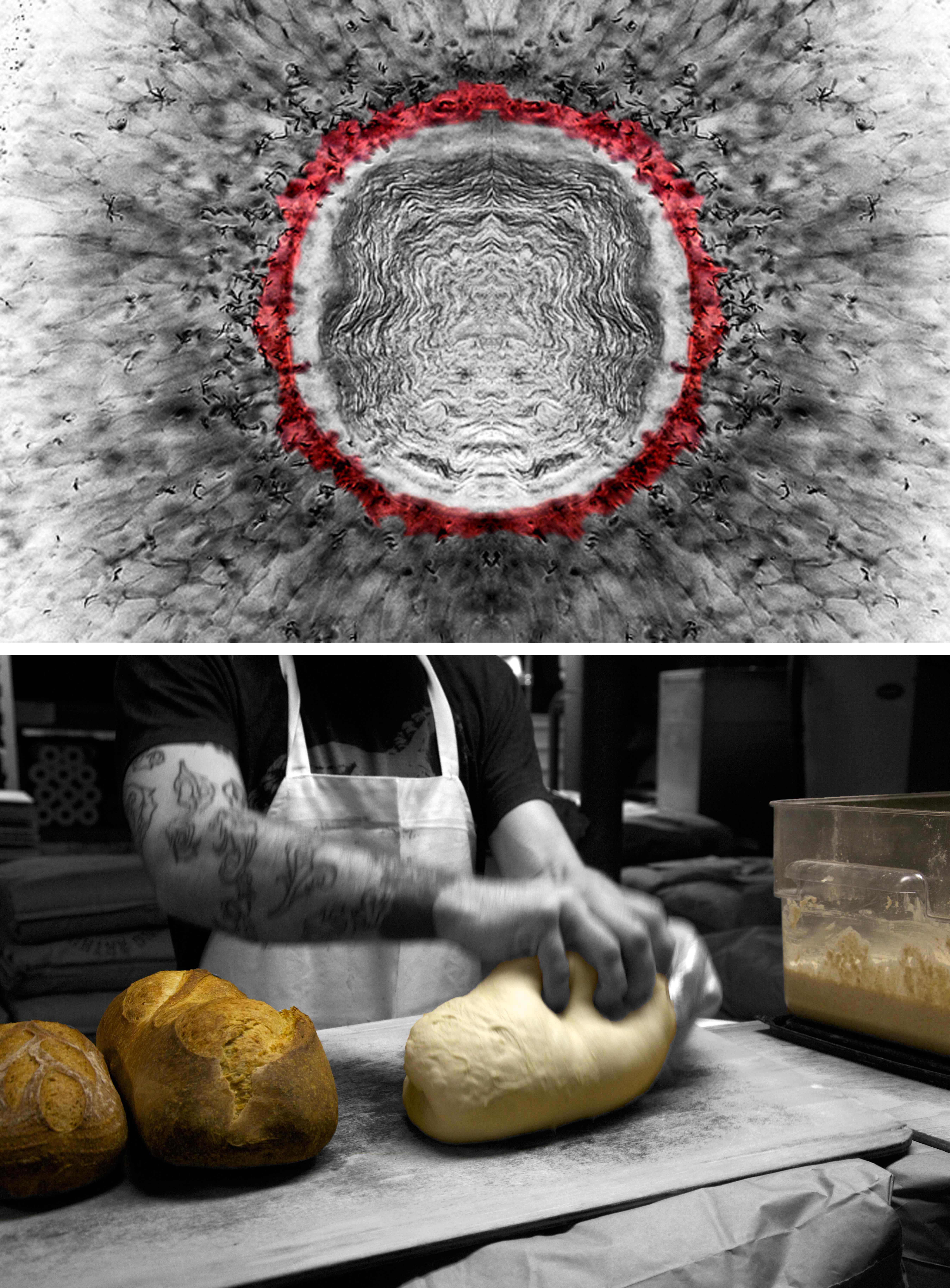
Figure 2- Stem cells and their niche. Upon division of a stem cell, one daughter will maintain the stem cell fate, and the other will produce a differentiated progeny. Stem cells are positioned in a restricted spatial niche, providing signals that maintain them in the proliferative, non-differentiated state. Following division, only one progeny is maintained in the niche, whereas the other will move out of the niche and differentiate. Top: once the eye of the zebra fish is specified, cells differentiate and produce neurons that sense light. Retinal stem cells (red) are maintained throughout the animal’s life in a niche, located adjacent to the lens (K. Cerveny and S. Wilson, University College London). Bottom: the live yeast stock termed ‘mother’ is carefully maintained in the bakery. Portions are allocated to produce dough and bread. A fine balance is kept in order to ensure a steady production of bread, while propagating the yeast stock (B. Shilo, Hi Rise Bakery, Cambridge, MA, USA).
Given my long-standing interest in photography, I decided to capture these images myself. As with the scientific images, the colorful pictures of the human world may contain too many hues that divert the viewer’s attention from the intended point. Therefore, I converted these pictures to black and white format as well, leaving only the topic of interest in color. By generating a common format for each image pair, the shared concept is visually reinforced. After assembling 35 pairs of images that cover the main concepts I delineated, the project was ready for presentation.
I have developed several means by which the project can be displayed. First, the images can be presented as a photography exhibit (Fig. 3), with a short introduction and a paragraph explaining the concept behind each image pair displayed next to the pictures. The images follow a scientifically logical order, starting with the pathways by which cells communicate during development, and how these allow elaborate patterns to be created. The exhibit then displays the concept of morphogens and evolution of patterns during speciation, followed by the execution of patterning that leads to the formation of tissues and organs. The exhibit ends with stem cells and the promise of induced pluripotent stem cells, and their possible utilization to create new organs. This exhibition has been displayed at the Radcliffe gallery at Harvard, at the Harvard educational portal at Allston (MA, USA) and at the Weizmann Institute of Science in Israel. I was concerned that the short explanations may not be sufficient to draw the audience into the subject matter, in the absence of a deeper explanation or a gallery talk. However, the audience response was extremely rewarding, and the best indication for the success of the approach was that people came to me with new suggestions for metaphors, indicating that they understood the concepts and were able to relate them creatively to their own world.
This work was also presented in the format of a public lecture, using the images as a way to illustrate and present in greater depth concepts of developmental biology. The fact that the storyline goes back and forth between scientific aspects, which may be more abstract, to images from daily life, which are readily appealing, keeps the audience alert and attracted to the lecture’s thread. Over the past year, I have given this lecture numerous times, in different continents and to very diverse audiences, from a lay audience including students and teachers, to scientists from different disciplines [see, for example, the lecture given at the European Molecular Biology Laboratory in 2012 (http://medias01-web.embl.de/Mediasite/Play/102f090bfa93473eb369a8c646a8d09c1d)]. The presentation proved to be highly effective for all audiences, again stimulating suggestions for new metaphors from the participants. At the outset, the most unpredictable audience was that of experts in developmental biology, as I was worried that this mode of presentation may seem trivial or over simplified. Here, I was pleasantly surprised, as the process of crystallizing complicated concepts to a single metaphor proved to be thought provoking, even for specialists.

Figure 3- Exhibit at the Harvard Allston Education Portal, April 2012. Photo: B. Shilo.
Finally, I have written a popular book that will be published by Yale University Press in the coming year. The text of this book flows in a manner that is independent of the images, but hopefully the accompanying pictures will make the book more accessible and stimulating. The book is aimed first and foremost at people with little or no background in developmental biology who are curious to learn about embryonic development. Although not a textbook, it could also be used as a supplement to developmental biology courses at high school or college undergraduate level. In addition, it may stimulate students to photograph their own metaphors to the developmental concepts they study, and a web forum could be constructed to display and discuss these analogies. Finally, students who begin their research in developmental biology labs are initially swamped by literature related to their future research project. It may be instrumental and inspiring if in parallel they could also read a book that provides them with a ‘bird’s eye’ view of the field and the emerging concepts, to place their particular project in the context of the broader picture.
Taking a wider view, one can ask if this approach of using human metaphors can be extended further, and used to present other scientific topics and disciplines. I found the analogy between a cell and a human highly effective, giving a clear and consistent definition of the individual unit. One can easily envisage extending this approach to fields such as cellular immunology or cancer biology. It may become more challenging to define the ‘quantum’ entity that would be analogous to humans, when considering other scientific fields and disciplines. I am certain, however, that creative modes could be devised to extend analogies to the human world in new ways, in order to enhance the intuitive understanding of complex scientific principles in a broad range of disciplines.
Ben-Zion Shilo, Weizmann Institute of Science, Israel
Follow Benny’s lead and enter our outreach competition by finding your own science metaphor!
 (2 votes)
(2 votes)
 Loading...
Loading...
 This post is part of a series on science outreach. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on science outreach. You can read the introduction to the series here and read other posts in this series here.


 (2 votes)
(2 votes)

 (No Ratings Yet)
(No Ratings Yet)
 Terminally differentiated cells are generally considered to be in a developmentally locked state in vivo; they are incapable of being directly reprogrammed into an entirely different state. Now, on
Terminally differentiated cells are generally considered to be in a developmentally locked state in vivo; they are incapable of being directly reprogrammed into an entirely different state. Now, on  Zebrafish have a remarkable capacity to regenerate and, as such, are being used increasingly to study stem cells and organ regeneration. Here, Chen-Hui Chen, Kenneth Poss and colleagues establish a luciferase-based approach for visualising stem cells and regeneration in adult zebrafish (
Zebrafish have a remarkable capacity to regenerate and, as such, are being used increasingly to study stem cells and organ regeneration. Here, Chen-Hui Chen, Kenneth Poss and colleagues establish a luciferase-based approach for visualising stem cells and regeneration in adult zebrafish ( Wnt signalling plays important roles during embryonic patterning and in tissue homeostasis, and mutations that affect the Wnt pathway are associated with cancer. Despite this, the exact way in which the Wnt pathway is regulated is still not fully understood. Now, Amy Bejsovec and colleagues uncover a novel regulator of Wnt signalling (
Wnt signalling plays important roles during embryonic patterning and in tissue homeostasis, and mutations that affect the Wnt pathway are associated with cancer. Despite this, the exact way in which the Wnt pathway is regulated is still not fully understood. Now, Amy Bejsovec and colleagues uncover a novel regulator of Wnt signalling ( The hair follicle epithelium forms a tube-like structure that is continuous with the epidermis, but how the lumen of this structure is created during morphogenesis and regeneration remains unclear. Now, Sunny Wong and colleagues identify a novel population of cells that initiates hair follicle lumen formation in mice (
The hair follicle epithelium forms a tube-like structure that is continuous with the epidermis, but how the lumen of this structure is created during morphogenesis and regeneration remains unclear. Now, Sunny Wong and colleagues identify a novel population of cells that initiates hair follicle lumen formation in mice ( The pituitary gland is an endocrine organ that plays a role in various physiological processes, including growth, metabolism and reproduction. The development of various pituitary endocrine cells is influenced by a number of transcription factors and signals. In this issue (
The pituitary gland is an endocrine organ that plays a role in various physiological processes, including growth, metabolism and reproduction. The development of various pituitary endocrine cells is influenced by a number of transcription factors and signals. In this issue ( The neural crest (NC) is a transient structure that gives rise to multiple lineages. Despite intense studies, it is still unclear whether the NC represents a homogeneous population of cells. Here, Jean Paul Thiery and colleagues examine this issue (
The neural crest (NC) is a transient structure that gives rise to multiple lineages. Despite intense studies, it is still unclear whether the NC represents a homogeneous population of cells. Here, Jean Paul Thiery and colleagues examine this issue ( Many organisms and their constituent tissues and organs vary substantially in size but differ little in morphology; they appear to be scaled versions of a common template or pattern. Here, David Umulis and Hans Othmer investigate the underlying principles needed for understanding the mechanisms that can produce scale invariance in spatial pattern formation and discuss examples of systems that scale during development. See the Review article on p.
Many organisms and their constituent tissues and organs vary substantially in size but differ little in morphology; they appear to be scaled versions of a common template or pattern. Here, David Umulis and Hans Othmer investigate the underlying principles needed for understanding the mechanisms that can produce scale invariance in spatial pattern formation and discuss examples of systems that scale during development. See the Review article on p.  How can the revolution in our understanding of embryonic development and stem cells be conveyed to the general public? Here, Ben-Zion Shilo presents a photographic approach to highlight scientific concepts of pattern formation using metaphors from daily life, displaying pairs of images of embryonic development and the corresponding human analogy. See the Spotlight article on p.
How can the revolution in our understanding of embryonic development and stem cells be conveyed to the general public? Here, Ben-Zion Shilo presents a photographic approach to highlight scientific concepts of pattern formation using metaphors from daily life, displaying pairs of images of embryonic development and the corresponding human analogy. See the Spotlight article on p. 



 – Atsushi Miyawaki and colleagues wrote about their recent paper in Development using
– Atsushi Miyawaki and colleagues wrote about their recent paper in Development using 
