December in preprints
Posted by the Node, on 19 January 2026
Welcome to our monthly trawl for developmental and stem cell biology (and related) preprints.
The preprints this month are hosted on bioRxiv – use these links below to get to the section you want:
- Patterning & signalling
- Morphogenesis & mechanics
- Genes & genomes
- Stem cells, regeneration & disease modelling
- Plant development
- Environment, evolution and development
Research practice and education
Spotted a preprint in this list that you love? If you’re keen to gain some science writing experience and be part of a friendly, diverse and international community, consider joining preLights and writing a preprint highlight article.
Developmental biology
| Patterning & signalling
Smarcc1 is essential for the patterning of the optic stalk and differentiation of the optic nerve head astrocytes
Nitay Zuk-Bar, Shai Ovadia, Guizhong Cui, Alexey Obolensky, Eyal Banin, Ron Ofri, Naihe Jing, Ruth Ashery-Padan
An NKX2-5 homolog is required downstream of BMP signaling to pattern the sensory-adhesive organ of a tunicate larva
Christopher J. Johnson, Joshua Kavaler, Christina D. Cota, Alberto Stolfi
PBX-dependent and independent Hox programs establish and maintain motor neuron terminal identity
Manasa Prahlad, Weidong Feng, Oyunsuvd Bat-Erdene, Yihan Chen, Paschalis Kratsios
The extracellular matrix in selective decussation of retinal ganglion cell axons: β2 laminins regulate the ipsilateral projection
Alanis Hernandez-Arce, Madeline Turo, Adam N. Robinson, Skylyn McNamara, Danny Yeo, Reyna I. Martínez-De Luna
EXPRESSION OF ANO1 IN HUMAN GASTROINTESTINAL TRACT DURING EMBRYONIC AND FETAL DEVELOPMENT
Vladimir Petrović, Aleksandra Veličkov, Marko Jović, Julija Radenković, Braca Kundalić, Dušan Miljković, Vukota Radovanović, Goran Radenković
Mon1‑Rab7 axis is essential for transport, localization and anchoring of oskar mRNA
Vasudha Dwivedi, Vrushali Katagade, Sourav Halder, Jyotish Sudhakaran, T Anjana, Girish S Ratnaparkhi, Vasudevan Seshadri, Anuradha Ratnaparkhi
Intrinsic and non-cell autonomous roles for a neurodevelopmental syndrome-linked transcription factor
Jayson J. Smith, Seth R. Taylor, Honorine Destain, Grace Kim, David H. Hall, John G. White, David M. Miller III, Paschalis Kratsios
Wnt and Nodal asymmetries stratify mouse laterality phenotypes in the absence of node flow
Amaia Ochandorena-Saa, Emeline Perthame, Zoé Oulerich, Alexander Chamolly, Thierry Blisnick, Johanna Lokmer, Cécile Rouillon, Philippe Bastin, Sigolène M. Meilhac
Comparison between the activities of canonical Wnt ligands in human pluripotent stem cell differentiation
Eleni Anastasia Rizou, Aryeh Warmflash
Fetoplacental circadian rhythms develop and then synchronize to the mother in utero
K.L. Nikhil, Keenan Bates, Elizabeth Sapiro, Jacob L. Amme, Ronald McCarthy, Sarah L. Speck, Varun Vasireddy, Ethan Roberts, Carmel A. Martin-Fairey, Miguel-E. Domínguez-Romero, Sandra Paola Cárdenas-García, Sarah K. England, Erik D. Herzog
Regulation of motor neuron differentiation in the Ciona larva
Sydney Popsuj, Tenzin Kalsang, Christina D. Cota, Alberto Stolfi
Notch receptors involved in the choice between intestinal secretory and enterocytes and differentiation of Bestrophin 4 cells
Samah Allayati, Pijush Sutradhar, Morgan Prochaska, Lea Maney, Christian Choy, Abrielle Swartz, Kenneth Wallace
Developmental regulation of intestinal best4+ cells
Abhinav Sur, Ella X. Segal, Michael P. Nunneley, Jason W. Sinclair, Morgan Kathleen Prochaska, Louis E. Dye, Yalan Wu, Liezhen Fu, Yun-Bo Shi, James Iben, Benjamin Feldman, Jeffrey A. Farrell
Scaling of the Bicoid morphogen gradient: the effect of state dependent diffusion
Priya Chakraborty, Shyam Iyer, Richa Rikhy, Mithun K. Mitra, Amitabha Nandi
Retinoic acid coordinates the orderly construction of the mammalian body in the anterior-to-posterior sequence
Anita Banerjee, Sameera Krishna Yallapragada, Gabriel Torregrosa-Cortés, Bhakti J Vyas, Ramkumar Sambasivan
Optogenetic Rescue Reveals Spatiotemporal Rules of Germ-Layer Patterning
Naomi Baxter, Robert Piscopio, Joseph Rufo, Dasol Han, Isobel Whitehead, Jasmine Dhillon, Siddharth S. Dey, Maxwell Z. Wilson
Exosome secretion is required for sonic hedgehog dispersal and signal gradient formation in the embryonic limb mesenchyme
Sean Corcoran, Joshua Fisher, Timothy A. Sanders, Edwin Munro
Actin and myosin dynamics during epithelial remodeling in avian gastrulation
Yu Ieda, Carole Phan, Olinda Alegria-Prévot, Aurélien Villedieu, Jérôme Gros
α-Parvin regulation of cell re-arrangement is critical for ureteric bud branching morphogenesis
Xinyu Dong, Fabian Bock, Ali Hashmi, Nada Bulus, Glenda Mernaugh, Gema Bolas, Shensen Li, Wanying Zhu, Meiling Melzer, Kyle Brown, Colton Miller, Olga Viquéz, Eloi Montañez, Ambra Pozzi, Sara A. Wickström, Roy Zent
Mitophagy upregulates WNT5A/Ca2+ signalling to accelerate fibroblast migration and wound healing
Matthew Hunt, Monica Torres, Nuoqi Wang, Shannon Hinch, Margarita Chatzopoulou, Gustavo Urbano-Quispe, Etty Bachar-Wikström, Jakob D Wikström
Reciprocal interactions between EMT and BMP signalling drive collective cell invasion
Yuri Takahashi, Alexandra Neaverson, Lara Busby, Filip Twarowski, Carlos Camacho-Macorra, Guillermo Serrano Nájera, Benjamin Steventon
Temporal Control of Decidual Inflammation by HOXA10 is Essential for Implantation and its Dysregulation is Associated with Early Pregnancy Loss
R Sharma, B Negi, R Ponsankaran, S Patil, G Godbole, A Mishra, S Shyamal, D Modi
Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates sugar homeostasis in Drosophila
Arnab Karmakar, Padinjat Raghu
YAP levels regulate anteroposterior elongation of hESC-derived gastruloids
Elizabeth Abraham, Thomas Roule, Olivia Mae Pericak, Mikel Zubillaga, Naiara Akizu, Conchi Estaras
| Morphogenesis & mechanics
Characterizing the role of mitochondrial dynamics during Drosophila convergent extension using NADH fluorescence lifetime imaging
Maria Espana-Pena, Alan Woessner, Colten Nichols, Kyle P. Quinn, Adam C. Paré
Primordial cardiomyocytes orchestrate myocardial morphogenesis and vascularization but are dispensable for regeneration
Jisheng Sun, Lu Chen, Jinhu Wang
Murine implantation chamber formation precedes natural and artificial decidualization
Harini Raghu Kumar, Noura Massri, Aishwarya V Bhurke, Akanksha Kapur, Pooja Gadhiya, Ripla Arora
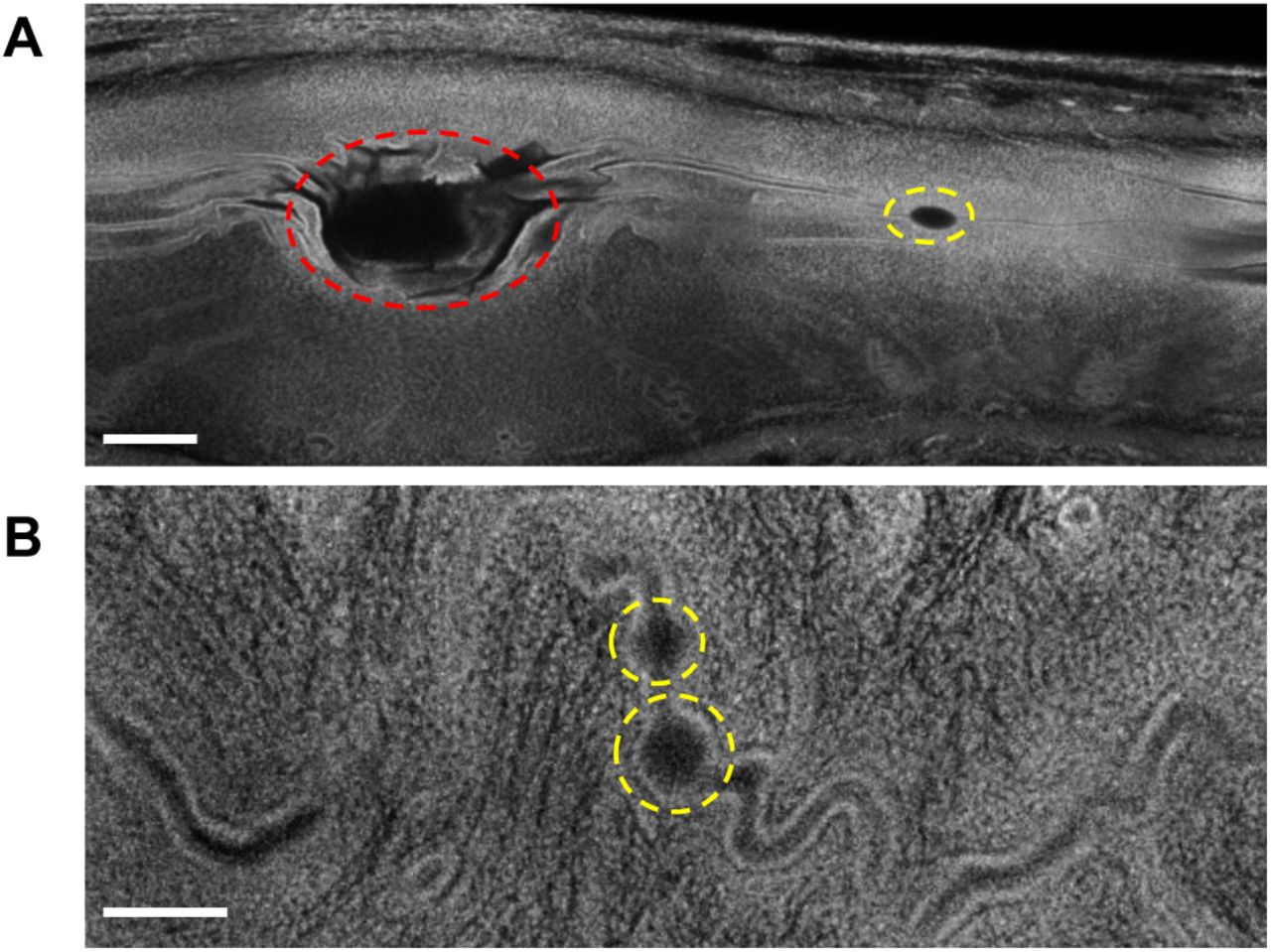
Genetically engineered ESC-derived embryos reveal Vinculin-dependent force responses required for mammalian neural tube closure
Ian S. Prudhomme, Eric R. Brooks, Nilay Taneja, Bhaswati Bhattacharya, Brian J. LaFleche, Yasuhide Furuta, Jennifer A. Zallen
Peristaltic contractions drive gut anisotropic growth through collective cell rearrangements
Koji Kawamura, Yoshiko Takahashi, Masafumi Inaba
HIF1α controls somitogenesis and spine development by regulating levels of intracellular oxygen in the presomitic mesoderm
Matthew J. Anderson, Angela Yao, Brittany Laslow, Ernestina Schipani, Mark Lewandoski
VEGF/ERK activation and PI3K inhibition together drive a vein-to-artery transition in an in vitro model of human angiogenesis
Amir Ugokwe, A.L. Pyke, E. Trimm, M. Chakraborty, X. Fan, L.T. Ang, K.M. Loh, K. Red-Horse
From patterning to secretion: Kv2.1 subunits as regulators of zebrafish hatching gland morphogenesis and function
Ruchi P Jain, Rosa R Amini, Vladimir Korzh
The developing tendon and enthesis are hypoxic and rely on hypoxia-inducible factor 1a (Hif1a) during postnatal development
Stephanie S. Steltzer, Nicole Migotsky, Tessa Phillips, Syeda N. Lamia, Ki Won Lee, Sueng-Ho Bae, Connor Leek, Sydney Grossman, Moaid Shaik, Allison Risha, Kaitlyn Frey, Claudia Loebel, Jun Hee Lee, Yatrik M. Shah, Adam C. Abraham, Megan L. Killian
Rootletin Fiber Dynamics Integrate Cytoskeletal Programs to Shape Neuroepithelial Architecture
Axelle Wilmerding, Glòria Casas Gimeno, Paula Espana-Bonilla, Susana Usieto, Murielle Saade
Cytoplasmatic polyadenylation of mRNA by TENT5A is critical for enamel mineralization
Goretti Aranaz-Novaliches, Olga Gewartowska, Frantisek Spoutil, Seweryn Mroczek, Pavel Talacko, Karel Harant, Ana-Matilde Augusto-Vale, Irena Krejzova, Carlos Eduardo Madureira Trufen, Pawel Krawczyk, Ales Benda, Vendula Novosadová, Radislav Sedlacek, Andrzej Dziembowski, Jan Prochazka
Loss of cilia drives centriole clustering and elimination during mammalian spermatogenesis
Jun Jie Chen, Xiangyu Gong, Michael Mak, Feng-Qian Li, Ken-Ichi Takemaru
Maturation Differentially Regulate Protein Kinase C-Mediated BK Channel Activation in Ovine Middle Cerebral Artery
Michell Goyal, Ravi Goyal
Mitophagy promotes metabolic reprogramming to enhance keratinocyte migration via ANGPTL4 during wound healing
Matthew Hunt, Nuoqi Wang, Monica Torres, Jenna Villman, Ilkka Paatero, Shannon Hinch, Gustavo Urbano-Quispe, Margarita Chatzopoulou, Etty Bachar-Wikström, Jakob D Wikström
Girdin controls the pace of 3D tracheal cell intercalation by coupling adherens junctions to the actin cytoskeleton in Drosophila
Sandra Carvalho, Patrick Laprise, Antoine Guichet, Véronique Brodu
| Genes & genomes
Hepatocyte-like cells die via steroid hormone and nuclear receptor E75-mediated apoptosis
Devika Radhakrishnan, Noah Landgraf, Luigi Zechini, Alessandro Scopelliti, Neha Agrawal
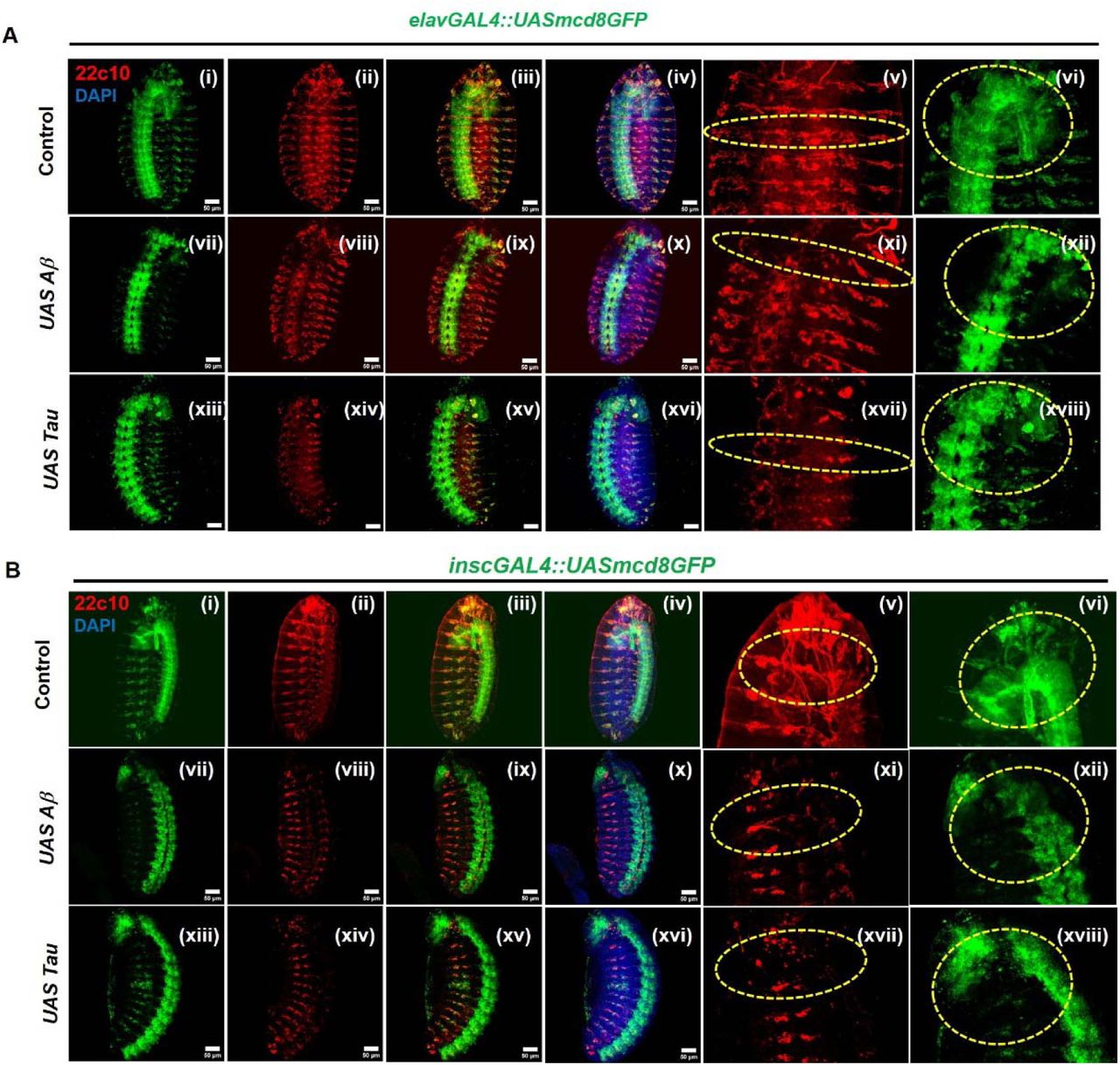
Early disruption of neurogenesis and neural architecture by Amyloid-β and Tau during Drosophila development
Khushboo Sharma, Neha Tiwari, Madhu G. Tapadia
GnRH-1 Neurons Are Not in the Goofy Group: 123cre Tracing Sets the Record Straight
Enrico Amato Jr., Mia V. Call, Noah M. LeFever, Mya Aviles-Carlos, Nikki M. Dolphin, Paolo E. Forni
Cysteine protease cathepsin B promotes high population density-induced mutagenesis, driving genome evolution and competitive growth in response to the crowding stress
Bin Yu, Yuji Suehiro, Bryan J. Johnson, Eui-Seung Lee, Dongdong Li, Yawen Huang, Joshua Johnson, Guangshuo Ou, James DeGregori, Shohei Mitani, Ding Xue
Endogenous retrovirus IAP forms virus-like particles and traffics across the maternal-fetal barrier
Abby J. Bergman, Guillaume Cornelis, Julie C. Baker
Functional architecture of cardiac TF regulatory landscapes in control of mammalian heart development
Virginia Roland, Johannes Tüchler, Andrea Esposito, Mattia Conte, Matteo Zoia, Ekapaksi Wisnumurti, Virginie Tissières, Julie Gamart, Raquel Rouco Garcia, Ines J. Marques, Akshay Akshay, Vincent Rapp, Brandon J. Mannion, Jennifer A. Akiyama, Prateek Arora, Harry Walker, Ali Hashemi Gheinani, Beth A. Firulli, Gretel Nusspaumer, Anthony B. Firulli, Guillaume Andrey, Axel Visel, Nadia Mercader, Javier Lopez-Rios, Mario Nicodemi, Iros Barozzi, Marco Osterwalder
Gadd45 regulates fate decisions of myeloid-type blood progenitor cells in Drosophila
Priyasi Jaiswal, Bama Charan Mondal
MYC/MAX balance dictates cell progenitor fate by altering the HOX program in the Drosophila eye
Sara Monticelli, Giorgio Milazzo, Suleman Khan Zadran, Martina Santulli, Nicola Balboni, Silvia Strocchi, Ettore De Giorgio, Pieter Mestdagh, Angela Giangrande, Roberto Bernardoni, Giovanni Perini
Inferring Cell Differentiation Dynamics with Unobserved Progenitors
William Howard-Snyder, Richard Zhang, Henri Schmidt, Michelle Chan, Benjamin J. Raphael
Transcriptional control of neuronal maintenance by SOX2 during inner ear innervation
Sukanya Raman, Akshara Dubey, Anubhav Prakash, Raman Kaushik, Lakshini Kannan, Palak Chugh, Raj K Ladher
Genetic characterization of the apterous Life Span Enhancer in Drosophila melanogaster
Cindy Reinger, Michèle Sickmann, Dimitri Bieli, Klemens E. Fröhlich, Alexander Schmidt, Markus Affolter, Martin Müller
Modular to cyclic TCA governs hematopoiesis in Drosophila
Ajay Tomar, Shaon Chakrabarti, Tina Mukherjee
Chromatin Accessibility Shapes Developmental-Specific Lineage Plasticity in Hematopoiesis
Sara Palo, Keiki Nagaharu, Mikael Sommarin, Rasmus Olofzon, Virginia Turati, Shamit Soneji, Göran Karlsson, Charlotta Böiers
| Stem cells, regeneration & disease modelling
FTO promotes skeletal muscle differentiation and regeneration by regulating m6A-modified c-Myc
Paromita Dey, Bijan K. Dey
Activin/TGF-beta signaling levels coordinate whole-body regeneration with genotoxic stress in Schmidtea mediterranea
Haleigh Brownlee, Amit Dubey, Nirurita Mahadev, Zachary Castles, Andrea Rauschmayer, Hannah Ashraf, Blair W. Benham-Pyle
Wnt/β-catenin signaling promotes zebrafish osteoblast dedifferentiation by wnt10a-mediated inhibition of NF-κB
Hossein Falah Mohammadi, Denise Posadas Pena, Dila Gülensoy, Ivonne Sehring, Gilbert Weidinger
Sustained ERK signaling couples the injury response to organizer formation during Hydra head regeneration
I.Y. Juanico, A.W. Stockinger, A.K. Virgen, N. Srisrimal, S.E. Campos, C.E. Juliano
Seizures, increased interhemispheric synchrony, altered brain transcriptomics and a leaky blood-brain barrier result from loss of ap3b2 in a CRISPR tadpole model of DEE48
Sulagna Banerjee, Cabriana W. Earl, Samuel C. Robson, Paul Szyszka, Caroline W. Beck
Metabolic Maturation Unveils Left Ventricular Identity in WNT ON/OFF Human Pluripotent Stem Cell-Derived Cardiomyocytes
Joaquín Smucler, Julia María Halek, Denisse Saulnier, Sheila Lucia Castañeda, Agustina Scarafía, Guadalupe Amín, Alejandra Guberman, Gustavo Sevlever, Santiago Miriuka, Lucía Natalia Moro, Ariel Waisman
A stem cell knockout village reveals lineage rewiring and a non-canonical islet cell fate in monogenic diabetes
Dingyu Liu, Bicna Song, Zhaoheng Li, Stephen Zhang, Tabassum Fabiha, Jiahui Zhao, Ayaka Inoki, Julie Piccand, Chew-Li Soh, Gary Dixon, Aaron Zhong, Nan Hu, Renhe Luo, Batu Ozlusen, Vipin Menon, Ting Zhou, Xiaojie Qiu, Gerard Gradwohl, Dapeng Yang, Kushal Dey, Wei Sun, Wei Li, Danwei Huangfu
Human embryo implantation involves Syncytin-2/MFSD2A-mediated heterokaryon formation with maternal endometrium
Tomas E. J. C. Noordzij, Martina Celotti, Ruben van Esch, Lisa Sackmann, Adriana Martìnez-Silgado, Franka de Jong, Hiromune Eto, Harry Begthel, Jeroen Korving, Theresa M. Sommer, Gaby S. Steba, Nicolas Rivron, Esther B. Baart, Johan H. van Es, Hans Clevers, Katharina F. Sonnen
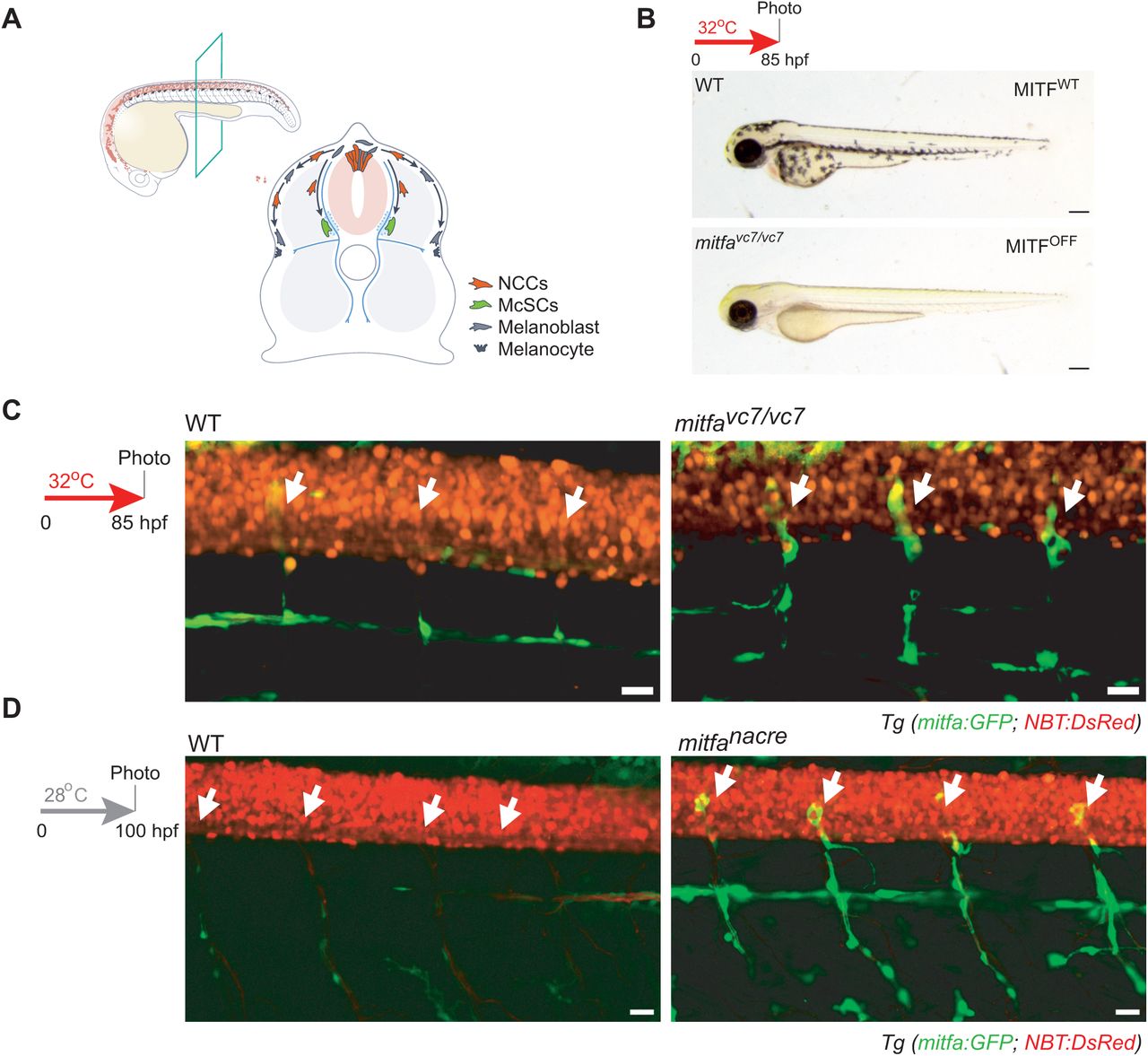
Loss of MITF activity leads to emergent cell states from the melanocyte stem cell lineage
Alessandro Brombin, Stephanie MacMaster, Jana Travnickova, Cameron Wyatt, Hannah Brunsdon, Emma Ramsey, Hong Nhung Vu, Eirikur Steingrimsson, Tamir Chandra, E. Elizabeth Patton
Ecdysone Receptor autonomously controls germ cell differentiation in the Drosophila ovary
Lauren E. Jung, Alexandria I. Warren, Changhong Yin, Weihua Huang, Allison C. Simmons, Samantha I. McDonald, Lindsay A. Swain, Victoria E. Garrido, Daniel N. Phipps, BiClaireline Cesar, Danielle S. Finger, Zhipeng Sun, Todd G. Nystul, Elizabeth T. Ables
Scube2 Modulates Coronary Vessel Formation during Cardiac Growth and Regeneration in Zebrafish
Ann Nee Lee, Ke-Hsuan Wei, Kaushik Chowdhury, Muhammad Abdul Rouf, An-Ju Chu, Yu-Jen Hung, Ku-Chi Tsao, Yan-Ting Chen, Yuh-Charn Lin, Yao-Ming Chang, Rubén Marín-Juez, Ruey-Bing Yang, Shih-Lei (Ben) Lai
Gestational inhibition of CSF1R signaling using PLX5622 drives musculoskeletal changes in postnatal offspring
Rouzbeh Ostadsharif Memar, Matthew Rosin, Siddharth R. Vora, Jessica M. Rosin
Placental insufficiency causes fetal growth restriction in mice lacking Delta-like homologue 1
Maria Lillina Vignola, Ruben Esse, Valeria Scagliotti, Chiara Servadei, Dominika Kardasz, Eugenia Marinelli, Claire Dent, Marika Charalambous
Spatial gene expression maps in vertebrate limbs display conserved and regenerative species-specific features within connective tissue
Conor L. McMann, Chanyoung Park, Jennifer K. Cloutier, Peter W. Reddien
Human-specific NOTCH2NL promotes astrogenesis by expanding proliferative glial progenitor states
Riina Ishiwatari, Xuanhao D. Sheu, Rintaro Amano, Yuki Y. Yamauchi, Pauline Rouillard, Takuma Kumamoto, Yusuke Kishi, Kazuo Emoto, Ikuo K. Suzuki
Proteostasis Remodeling Across Development Defines Fetal, Neonatal, and Adult Hematopoietic Stem Cell States
Helena Yu, Yoon Joon Kim, Katelyn Chen, Andrea Z. Liu, Mary Jean Sunshine, Robert A.J. Signer
Mesodermal-niche interactions direct specification and differentiation of pancreatic islet cells in human multilineage organoids
Georgina Goss, Alejo Torres-Cano, Martina Pedna, Heather Wilson, Michelle Simon, Flavia Flaviani, Alessandra Vigilante, Francesca M. Spagnoli
A novel and critical role of the intracellular Zona Pellucida protein 2 (ZP2) for blastocyst formation in mice
Thomas Nolte, Steffen Israel, Hannes C.A. Drexler, Georg Fuellen, Michele Boiani
A Single Cell Atlas of the Newt Iris During Lens Regeneration
Olivia M. Williams, Kelsey E. Ahearn, Joseph L. Sevigny, Nicole Farber, Disha Hegde, Kenneth J. Lampel, Jenna Loporcaro, Leo Napoleon, Jacob Nipoti, Timothy Ralich, Brooklyn Wallace, W. Kelley Thomas, Konstantinos Sousounis
Mosaic hotspot PIK3CA mutations cause non-cell-autonomous vascular overgrowth and pan-lineage dysregulation at disease onset
Hannah Brunsdon, Nuoya Wang, Micha Sam Brickman Raredon, Ralitsa R Madsen, Robert K Semple, E. Elizabeth Patton
AAV-mediated neuronal expression of FOXG1 restores oligodendrocyte maturation, myelination, and hippocampal structure in mouse models of FOXG1 syndrome
Jaein Park, Holly O’Shea, Shin Jeon, Dongjun Shin, Liwen Li, Seon Ung Hwang, Michael Kofi Anyane-Yeboa, Songlin Yang, Camille F. Harrison, Yeong Shin Yim, Jae W. Lee, Soo-Kyung Lee
The germline-restricted chromosome orchestrates germ cell development in passerine birds
Niki Vontzou, Yifan Pei, Israel Campo-Bes, Wolfgang Forstmeier, Moritz Hertel, Manuel Irimia, Bart Kempenaers, Sylvia Kuhn, Katrin Martin, Jakob C. Mueller, Kim Teltscher, Annelie Mollbrink, Xesús Abalo, Matthew T. Biegler, Simone Immler, Francisco J. Ruiz-Ruano, Alexander Suh
Rapamycin Differentially Impacts Germline Stem Cell Quiescence Across Diverse Genetic Backgrounds of Drosophila Melanogaster
Sahiti Peddibhotla, Miriam Gonzaga, Tricia Zhang, Yasha Goel, Jun Sun, Benjamin R. Harrison, Daniel E. L. Promislow, Hannele Ruohola-Baker
An RNA ligase shapes transcriptional profiles, neural function, and behaviour in the developing larval zebrafish
Fiona S. Klusmann, Anna C. Kögler, Katja Slangewal, Onur Önder, Heike Naumann, Andreas Marx, Armin Bahl, Patrick Müller
The regenerative potential of adult Nestin+ cerebellar astroglia is limited compared to in neonates
N. Sumru Bayin, Daniel N. Stephen, Richard Koche, Alexandra L. Joyner
| Plant development
Interplay between petal identity and cell layer identity in petunia flowers
Quentin Cavallini-Speisser, Emma Désert, Evelyne Duvernois-Berthet, Pierre Chambrier, Patrice Morel, Brice Letcher, Carine Rey, Jérémy Just, Suzanne Rodrigues Bento, Daniel Bouyer, Marie Monniaux
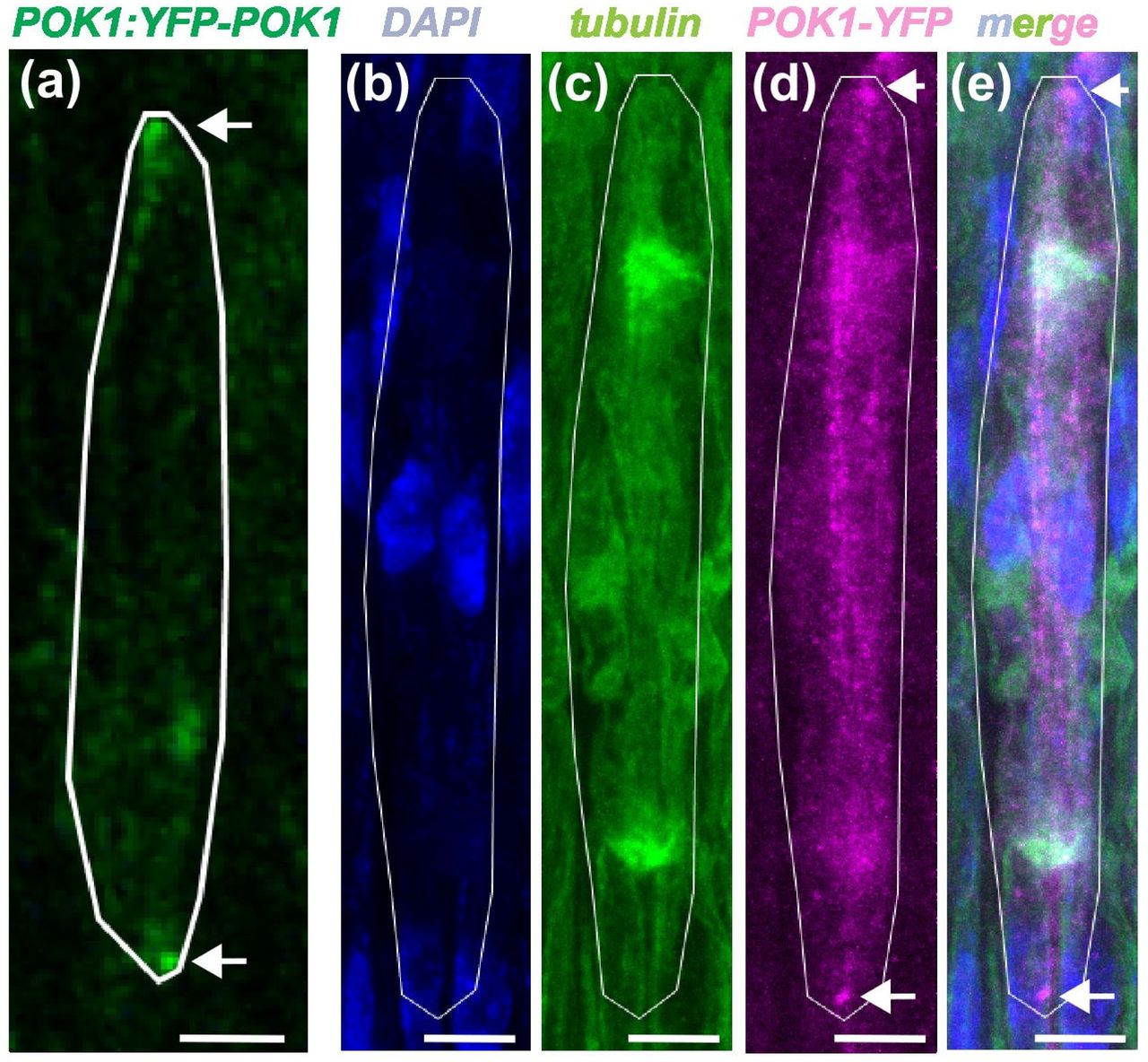
Robust division orientation of cambium stem cells requires cortical division zone components but not the preprophase band
Xiaomin Liu, Pantelis Livanos, Laura Sophie Schütz, Sabine Müller, Thomas Greb
Alternative splicing of PIF4 regulates plant development under heat stress
María Niño-González, Benjamin Alary, Dóra Szakonyi, Tom Laloum, Paula Duque, Guiomar Martín
The phytolongin AtPhyl2.1 is involved in cell plate formation and root development
Valerie Wattelet-Boyer, Matthieu Buridan, Yuri L. Negroni, Chiara Mafficini, Franziska Dittrich-Domergue, Lilly Maneta-Peyret, Emily Breeze, Michela Zottini, Elide Formentin, Francesco Filippini, Lysiane Brocard, Patrick Moreau
An embryo-derived peptide signal directs endosperm polarity in Arabidopsis
Audrey Creff, Jack Rhodes, Camille Salaün, Julien Larive, Vincent Bayle, Emma Turley, Tatsuya Nobori, Duarte D. Figueiredo, Benoit Landrein, Cyril Zipfel, Gwyneth Ingram
A SABATH family enzyme regulates development via the gibberellin-related pathway in the liverwort Marchantia polymorpha
Shogo Kawamura, Eita Shimokawa, Maika Ito, Isuzu Nakamura, Takehiko Kanazawa, Megumi Iwano, Rui Sun, Yoshihiro Yoshitake, Shohei Yamaoka, Shinjiro Yamaguchi, Takashi Ueda, Misako Kato, Takayuki Kohchi
Salt stress disrupts local auxin and COP1 gradients in Arabidopsis apical hooks
Elizabeth van Veen, Jesse J. Küpers, Xizheng Chen, Yu Him Tang, Thijs de Zeeuw, Kilian Duijts, Scott Hayes, Christa Testerink, Charlotte M. M. Gommers
CYSTEINE-RICH RLK2 regulates development via callose synthase-dependent symplastic transport in Arabidopsis
Adam Zeiner, Julia Krasensky-Wrzaczek, Sunita Jindal, Jakub Hajný, Mansi Sharma, Filis Morina, Elisa Andresen, Mirva Pääkkönen, Hendrik Küpper, Johannes Merilahti, Michael Wrzaczek
| Environment, evolution and development
Lateral inhibition governs ancestral cellular patterning in fossil and extant liverworts
Josep Mercadal, Susan Tremblay, Leonie Kraska, Martin A. Hutten, Pau Formosa-Jordan
Cell type diversification and phenotype convergence underlying white fin-ornamentation of cyprinid fishes
Delai Huang, Tiffany Liu, August A. Carr, Pietro H. de Mello, Yipeng Liang, Leah P. Shriver, François Chauvigné, Stephen L. Johnson, Joan Cerdà, Gary J. Patti, David M. Parichy
Epigenetic Coalitions Couple Tissue Growth to Generate Periodic Colour Patterns in Birds
Zhou Yu, Wei Zhao, Chih-Kuan Chen, Ya-Chen Liang, Hans I-Chen Harn, Wen-Chien Jea, Tzu-Yu Liu, Tsz Yau Law, Ting-Xin Jiang, Ping Wu, Edward Chuong, Qing Nie, Cheng-Ming Chuong
Spatial and temporal coordination of signaling pathways in tissue differentiation: developmental atlas of protein expression during zebra finch beak maturation
Renée A. Duckworth, Sarah E. Britton, Cody A. Lee, Kathryn C. Chenard, Alexander V. Badyaev
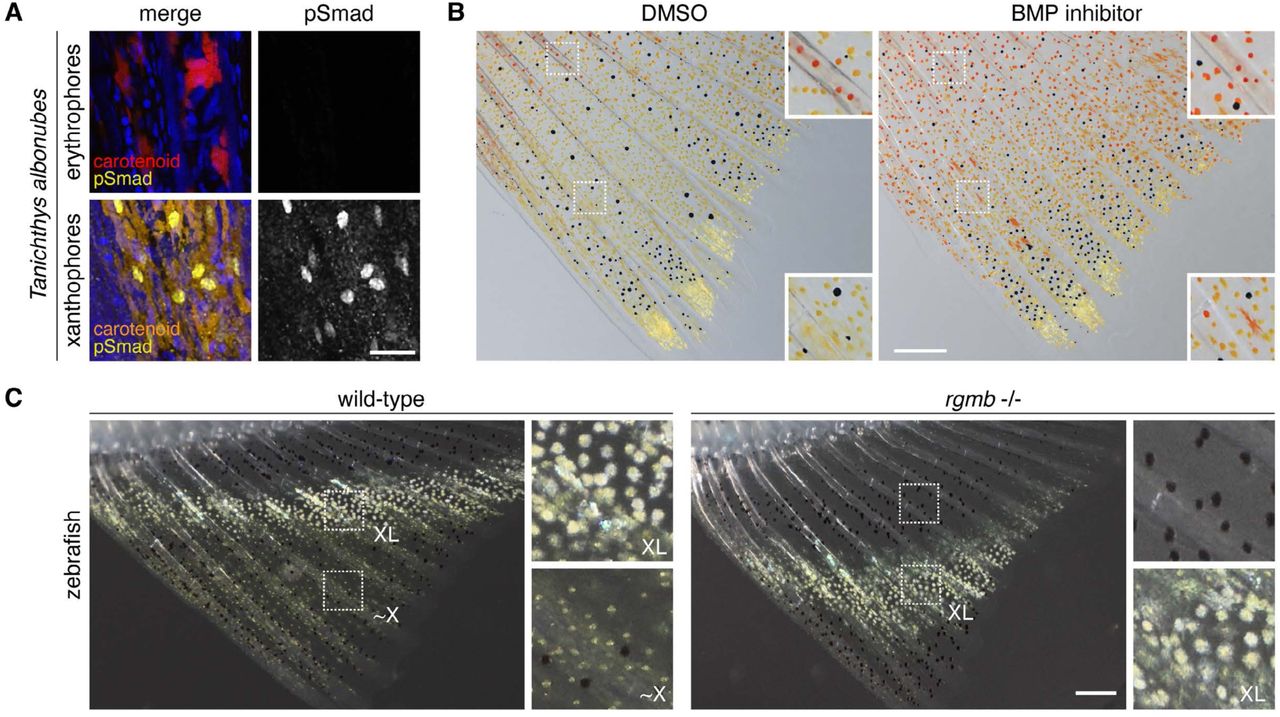
Graded BMP signals modulate yellow and red color in fishes impacting adult pigment pattern and behavior
Delai Huang, Pietro L. H. de Mello, Tiffany Liu, Yu Liu, Emaan H. Kapadia, Yipeng Liang, Jianguo Lu, Joseph C Corbo, David M Parichy
Embryonic development of the Mediterranean starfish Hacelia attenuata
Silvia Caballero-Mancebo, Laurent Gilletta, Janet Chenevert, Stefania Castagnetti
Multiple retinoic acid pathway factors function together during development of a mollusc
Kim Dao, J. David Lambert
Cell Biology
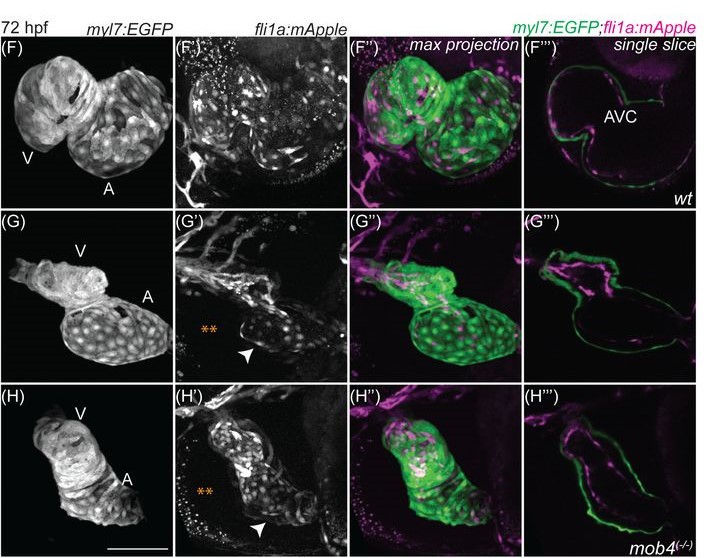
Role of the obligate STRIPAK complex component Mob4 in zebrafish vascular development and stability
Tvisha Misra, Shimon M. Rosenthal, Mengyi Song, Nathan J. Stutt, Laura McDonald, Ashish R. Deshwar, Anne-Claude Gingras, Ian C. Scott
Steroid hormone‑dependent glial‑neuronal interaction promotes brain development during Drosophila metamorphosis
Eisuke Imura, Naoki Okamoto, Naoki Yamanaka
3-Mercaptopyruvate Sulfurtransferase (MPST) Regulates Mitochondrial Metabolism and Epithelial Differentiation in Neonatal Patient-derived Airway Cells
Abhrajit Ganguly, Cynthia M. Carter, Aristides Rivera Negron, Paul T. Pierce, Lynette K. Rogers, Matthew S. Walters, Y.S. Prakash, Trent E. Tipple, Arlan Richardson
Development of the Early Childhood Duodenum across Ancestry, Geography and Environment
Joshua de Sousa Casal, Krishnan Raghunathan, Chelsea Asare, Abigail Plone, Nazanin Moradinasab, Junaid Iqbal, Lianna F. Wood, Elsy M. Ngwa, Xia Chen, S. Fisher Rhoads, Clara Baek, Dur-e Shahwar, Neha S. Dhaliwal, Madison Wong, Max Garrity-Janger, Lily P. Gillette, Stephanie Regis, Fatima Zulqarnain, Asra Usmani, Jason D. Boisvert, Casey R. Johnson, Jackson Larlee, Michael D. Anderson, Daniel Zeve, Elisa Saint-Denis, Thomas G. Wichman, Jeffrey La, Ashish Jain, Liang Sun, Lauren Scudari, Natalie N. Bhesania, Zehra Jamil, Michelle Galeas-Pena, Adam R. Greene, Aneeta Hotwani, Fedaa Najdawi, Shyam S. Raghavan, Donald E. Brown, Christopher A. Moskaluk, Heather H. Burris, Piotr Sliz, Phyllis R. Bishop, Scott B. Snapper, Kamran Sadiq, Sarah C. Glover, Muhammad Imran Nisar, Sana Syed, Jocelyn A. Silvester, Jose Ordovas-Montanes, Jay R. Thiagarajah
Oxygen availability and hypoxia-independent action of HIF1α controls human trophoblast maturation and function
Johanna Lattner, Javier Bregante, Michaela Burkon, Ornella Elezaj, Meritxell Huch, Michele Marass, Claudia Gerri
Constitutive Yap activation in distal nephron segments disrupts epithelial identity and nephron patterning
Zeinab Dehghani-Ghobadi, Eunah Chung, Mohammed Sayed, Christopher Ahn, Yueh-Chiang Hu, Hee-Woong Lim, Joo-Seop Park
Neonatal phlebotomy-induced anemia compromises mitochondrial bioenergetics in the developing hippocampus
Thomas W. Bastian, Diana J. Wallin, Amanda K. Barks, Raghavendra B Rao, Michael K. Georgieff
Proteomic profiling of Elp1-deficient trigeminal ganglia reveals disruption of neurotrophic and metabolic pathways in a familial dysautonomia mouse model
Carrie E. Leonard, Lauren Clarissa Tang, Beatrix Ueberheide, Lisa A. Taneyhill
aPKC-ζ III promotes trophoblast fusion by altering Par-3 interactions with Hippo Signaling Kinase LATS1
Sumaiyah Z. Shaha, Wendy K. Duan, Juan Garcia Rivas, Ivan K. Domingo, Meghan Riddell
Maternal protein restriction alters chromatin accessibility in neuroprogenitors of the fetal hypothalamus of rats
Valérie Amarger, Morgane Frapin, Pieter Vancamp
Human CSB-deficient iPSCs exhibit impaired DNA damage repair and stress responses following BPDE exposure in an early developmental model
Alessia Lofrano, Wasco Wruck, Nina Graffmann, James Adjaye
SMARCA2/4-Dependent Chromatin Remodelling Establishes Gene Regulatory Programs in Early Human Embryos and Blastoids
Sam S.F.A. van Knippenberg, Maria Tryfonos, Joke De Busscher, Mairim Solis, Chloe Lorent, Oceane Girard, Suresh Poovathingal, Marta Wojno, Sherif Khodeer, Jade De Clercq, Antonina Mikorska, Inge Smeers, Eva Wigerinck, Yara Meynen, Thierry Voet, Laurent David, Hilde Van de Velde, Vincent Pasque
The long noncoding RNA Peanut (Gm11454) promotes neurogenesis and rod photoreceptor differentiation during postnatal retinal development
Jade Enright Hostetler, Fion Shiau, Xiaodong Zhang, Shiming Chen, Philip A. Ruzycki, Seth Blackshaw, Brian S. Clark
Modelling
The (un)likelihood of clock-driven lateral root priming; A modeling exploration
Kirsten H. ten Tusscher
Cell size control emerges from the vein-dependent coordinated divisions of distinct cell groups in Drosophila wing
Kaoru Sugimura, Ryu Takayanagi, Toshinori Namba, Zeping Qu, Shuji Ishihara
Human trunk embryoids with patterned anterior-posterior and dorsal-ventral body axes: utility for understanding human development and disease
Tianming Wu, Hao Yu, Brian S.H. Wong, Kexin Teng, Weiman Xiang, Ling Xu, Jianan Zhang, Angel Y.F. Kam, Ethel S.K. Ng, Joaquim Vong, Jiannan Zhang, Bo Gao, Stephen K.W. Tsui, Stephen Dalton
Determining the age of single cells using scBayesAge
Chanyue Hu, Matteo Pellegrini
Cilia.io: Computer vision and machine learning reveal spatial patterns of cilia beating dynamics in the spinal cord
Ece Atayeter, Jason Ho, Talon G. Blottin, Ilyena B. Joe, Ron S. Sistrunk, Bo Zhang, Lilianna Solnica-Krezel, Andreas Gerstlauer, John B. Wallingford, Ryan S. Gray
Tools & Resources
Engineering protein expression dynamics with Tet-ON and dTAG degron systems: from precise control to oscillations
Benjamin Noble, Oliver Cottrell, Andrew Rowntree, Veronica Biga, Florence Woods, Xinjie Wang, Nancy Papalopulu, Anzy Miller
Integrated Single-cell Analysis Uncovers Regulatory Logic of Cranial Ectoderm Development
Ceren Pajanoja, Jenaid Rees, Ed Zandro M Taroc, Laura Kerosuo
Advancing Knock-In Approaches for Robust Genome Editing in Zebrafish
Anjelica Rodriguez-Parks, Ella Grace Beezley, Steffani Manna, Isabella Silaban, Sarah I Almutawa, Siyang Cao, Hossam Ahmed, Megan Guyer, Sean Baker, Mark P Richards, Junsu Kang
Spatiotemporal Atlas of Heart Development Reveals Blood-Flow-Dependent Cellular, Structural, Metabolic, and Spatial Remodeling
Jooyoung Park, Shuofei Sun, Rohit Agarwal, Andreas Stephanou, Mong Lung Steven Poon, Hyun Maeng, Peyton Lancaster, Iwijn De Vlaminck, Jonathan Butcher
The Heterogeneous Nature of Atrioventricular Conduction Tissues in Tetralogy of Fallot demonstrated by Hierarchical Phase-contrast Tomography
V Sabarigirivasan, J Brunet, H Dejea, A Crucean, A Jegatheeswaran, C Capelli, E Pajaziti, Theresa Urban, Joanna Purzycka, Paul Tafforeau, C L Walsh, P D Lee, A C Cook
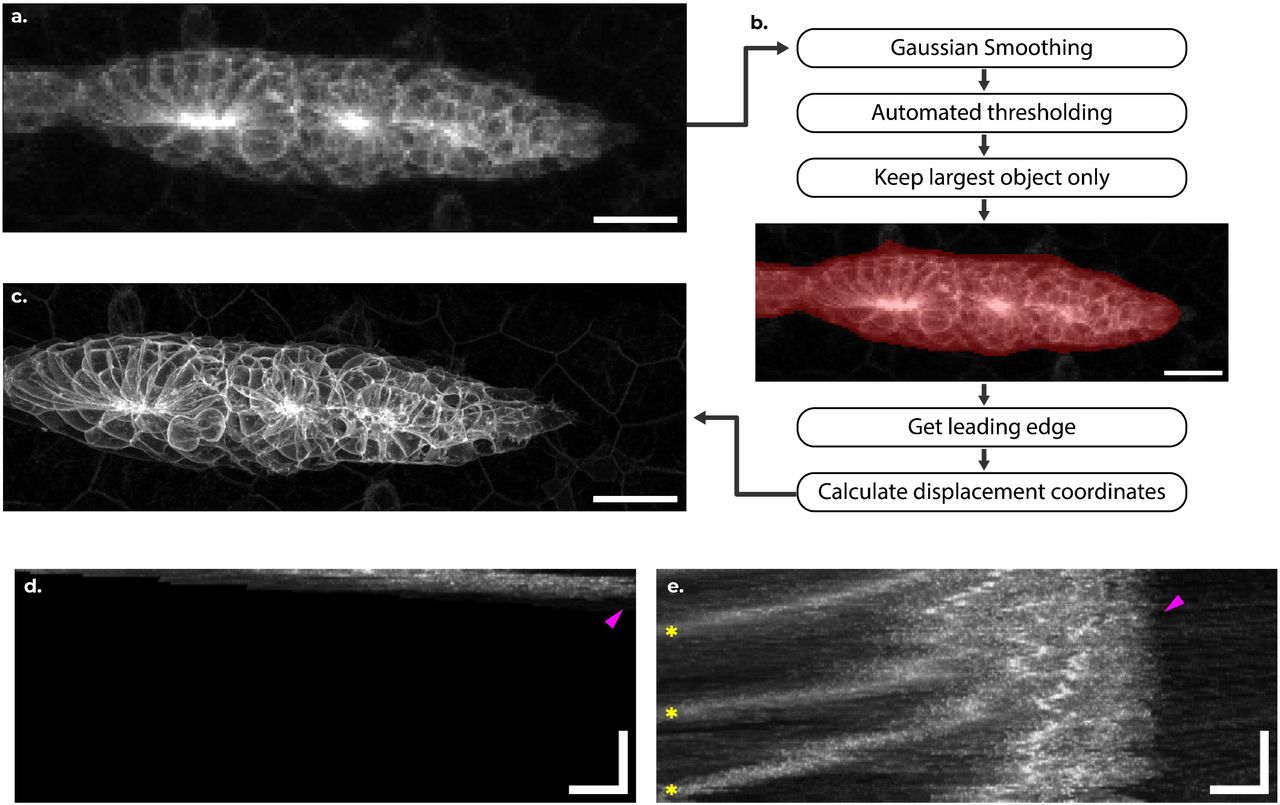
DySTrack: a modular smart microscopy tool for live tracking of dynamic samples on modern commercial microscopes
Zimeng Wu, Octavian Voiculescu, Alessandro Mongera, Roberto Mayor, Mie Wong, Jonas Hartmann
Assessing the impact of carrier solvent and solid phase extraction blank toxicity on fish embryo testing
Jakob Pfefferle, Sarah Johann, Henner Hollert, Riccardo Massei
Research practice & education
Chromatin profiling for everyone: FFPE-CUTAC for the theory and practice of modern molecular biology
Yiling Xu, Steven Henikoff, Kami Ahmad
The currency of research access: How undergraduates leverage social capital to gain research experience
Christopher James Zajic, Trevor T. Tuma, Erin L Dolan
Science-wide mapping and ranking of institutions based on affiliated authors
John P.A. Ioannidis, Jeroen Baas, Roy Boverhof, Cyril Voyant
A thirty-year trend of increasing clinical orientation at the National Institutes of Health
Brad L. Busse, James M. Tucker, Summer E. Allen, George M. Santangelo, Kristine A. Willis
Prediction of transformative breakthroughs in biomedical research
Matthew T. Davis, Brad L. Busse, Salsabil Arabi, Payam Meyer, Travis A. Hoppe, Rebecca A. Meseroll, B. Ian Hutchins, Kristine A. Willis, George M. Santangelo
Funders’ expectations for open science in cardiovascular research: A Scoping review of the largest cardiovascular research funders
Anna Catharina Vieira Armond, Al Mamoune Alaoui, David Moher, Jean Rouleau, Kelly D. Cobey
Using GPT-4 to Automate the Generation of Lay Summaries for Cancer Publications
Emma Purdie, Tony Yu, Jochen Weile, Diana Lemaire, Melanie Courtot
React-to-Me: A Conversational Interface for Interactive Exploration of the Reactome Pathway Knowledgebase
Helia Mohammadi, Fatemeh Almodaresi, Gregory F J Hogue, Adam Wright, Marija Orlic-Milacic, Nancy T Li, Amin Mawani, Lincoln Stein
To be or not to be a PI: Advice for trainees in the biomedical/basic sciences
Hei-Yong G. Lo, Lily L. Nguyen, Kristine M. Sikora, Bruce Mandt, Angeles B. Ribera
Trump Administration Impacts on Early Career Scientists and How To Fight Back
Crystal Hammond, John Patrick Flores, Siara Rouzer, Kassandra Fernandez, Amy Ralston, Adriana Bankston


 (No Ratings Yet)
(No Ratings Yet) (6 votes)
(6 votes)