March in preprints
Posted by the Node, on 8 April 2024
Welcome to our monthly trawl for developmental and stem cell biology (and related) preprints.
The preprints this month are hosted on bioRxiv and arXiv – use these links below to get to the section you want:
- Patterning & signalling
- Morphogenesis & mechanics
- Genes & genomes
- Stem cells, regeneration & disease modelling
- Plant development
- Evo-devo
Research practice and education
Developmental biology
| Patterning & signalling
Shruti S. Tophkhane, Sarah J. Gignac, Katherine Fu, Esther M. Verheyen, Joy M. Richman
Na Qu, Abdelkader Daoud, Daniel O. Kechele, Jorge O. Múnera
Two opposing roles for Bmp signalling in the development of electrosensory lateral line organs
Alexander S. Campbell, Martin Minařík, Roman Franěk, Michaela Vazačová, Miloš Havelka, David Gela, Martin Pšenička, Clare V. H. Baker
Zhilin Deng, Wenqi Chang, Chengni Li, Botong Li, Shuying Huang, Jingtong Huang, Ke Zhang, Yuanyuan Li, Xingdong Liu, Qin Ran, Zhenhua Guo, Sizhou Huang
Symmetry breaking and fate divergence during lateral inhibition in Drosophila
Minh-Son Phan, Jang-mi Kim, Cara Picciotto, Lydie Couturier, Nisha Veits, Khallil Mazouni, François Schweisguth
Gyunghee G. Lee, Aidan J. Peterson, Myung-Jun Kim, Michael B. O’Connor, Jae H. Park
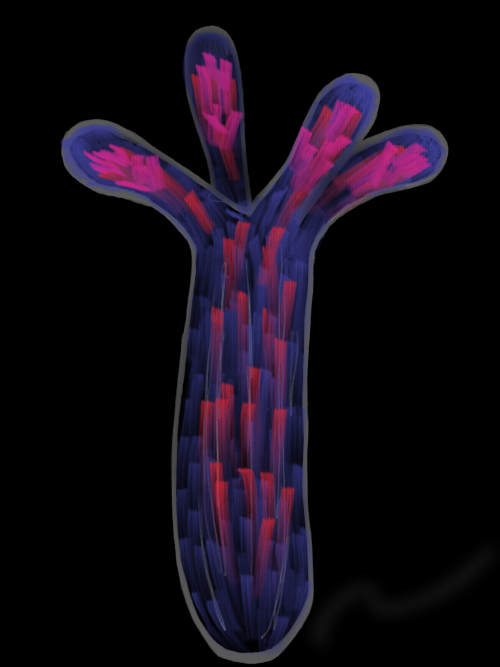
Dina Rekler, Shai Ofek, Sarah Kagan, Gilgi Friedlander, Chaya Kalcheim
A Foxf1-Wnt-Nr2f1 cascade promotes atrial cardiomyocyte differentiation in zebrafish
Ugo Coppola, Jennifer Kenney, Joshua S. Waxman
Vasileios R. Ouzounidis, Mattie Green, Charlotte de Ceuninck van Capelle, Clara Gebhardt, Helena Crellin, Cameron Finlayson, Bram Prevo, Dhanya K. Cheerambathur
Notch3 is a genetic modifier of NODAL signalling for patterning asymmetry during mouse heart looping
Tobias Holm Bønnelykke, Marie-Amandine Chabry, Emeline Perthame, Audrey Desgrange, Sigolène M. Meilhac
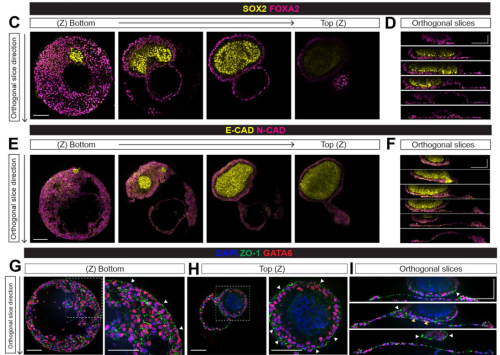
Miguel Angel Ortiz-Salazar, Elena Camacho-Aguilar, Aryeh Warmflash
| Morphogenesis & mechanics
Cerebellar granule cell migration and folia development requires Mllt11/Af1q/Tcf7c
Marley Blommers, Danielle Stanton-Turcotte, Emily A. Witt, Mohsen Heidari, Angelo Iulianella
Roles of TYRO3 Family Receptors in Germ Cell Development During Mouse Testis Formation
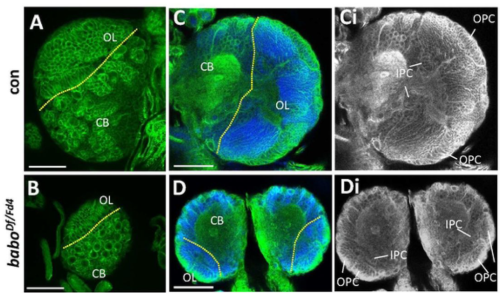
Zhenhua Ming, Stefan Bagheri-Fam, Emily R Frost, Janelle M Ryan, Michele D Binder, Vincent R Harley
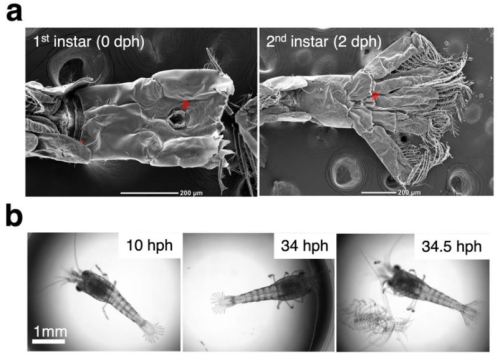
Post-embryonic tail development through molting of the freshwater shrimp Neocaridina denticulata
Haruhiko Adachi, Nobuko Moritoki, Tomoko Shindo, Kazuharu Arakawa
Hox-A2 protein expression in avian jaws cartilages and muscle primordia development
Stéphane Louryan, Myriam Duterre, Nathalie Vanmuylder
Estelle HIRSINGER, Cedrine BLAVET, Marie-Ange BONNIN, Lea BELLENGER, Tarek GHARSALLI, Delphine DUPREZ
Initiation and Formation of Stereocilia during the Development of Mouse Cochlear Hair Cells
Suraj Ranganath Chakravarthy, Thomas S. van Zanten, Raj K Ladher
Spns1-dependent endocardial lysosomal function drives valve morphogenesis through Notch1-signaling
Myra N. Chávez, Prateek Arora, Alexander Ernst, Marco Meer, Rodrigo A. Morales, Nadia Mercader
Development of Pial Collaterals by Extension of Pre-existing Artery Tips
Suraj Kumar, Niloufer Shanavas, Swarnadip Ghosh, Vinayak Sivaramakrishnan, Manish Dwari, Soumyashree Das
Kristen Kurtzeborn, Vladislav Iaroshenko, Tomáš Zárybnický, Julia Koivula, Heidi Anttonen, Darren Brigdewater, Ramaswamy Krishnan, Ping Chen, Satu Kuure
Venom gland organogenesis in the common house spider
Afrah Hassan, Grace Blakeley, Alistair P. McGregor, Giulia Zancolli
| Genes & genomes
José González-Martínez, Agustín Sánchez-Belmonte, Estefanía Ayala, Alejandro García, Enrique Nogueira, Jaime Muñoz, Anna Melati, Daniel Giménez, Ana Losada, Sagrario Ortega, Marcos Malumbres
Prabuddha Chakraborty, Terry Magnuson
Zhimin Xu, Zhao Wang, Lifang Wang, Yingchuan B. Qi
The transcriptomic landscape of monosomy X (45,X) during early human fetal and placental development
Jenifer P. Suntharalingham, Ignacio del Valle, Federica Buonocore, Sinead M. McGlacken-Byrne, Tony Brooks, Olumide K. Ogunbiyi, Danielle Liptrot, Nathan Dunton, Gaganjit K Madhan, Kate Metcalfe, Lydia Nel, Abigail R. Marshall, Miho Ishida, Neil J. Sebire, Gudrun E. Moore, Berta Crespo, Nita Solanky, Gerard S. Conway, John C. Achermann
Katherine L. Duval, Ashley R. Artis, Mary G. Goll
Temporally resolved early BMP-driven transcriptional cascade during human amnion specification
Nikola Sekulovski, Jenna C. Wettstein, Amber E. Carleton, Lauren N. Juga, Linnea E. Taniguchi, Xiaolong Ma, Sridhar Rao, Jenna K. Schmidt, Thaddeus G. Golos, Chien-Wei Lin, Kenichiro Taniguchi
Suchit Ahuja, Cynthia Adjekukor, Qing Li, Katrinka M. Kocha, Nicole Rosin, Elodie Labit, Sarthak Sinha, Ankita Narang, Quan Long, Jeff Biernaskie, Peng Huang, Sarah J. Childs
Screening of functional maternal-specific chromatin regulators in early embryonic development
Guifen Liu, Yiman Wang, Xiangxiu Wang, Wen Wang, Zheng Cao, Yong Zhang
Escape from X inactivation is directly modulated by levels of Xist non-coding RNA
Antonia Hauth, Jasper Panten, Emma Kneuss, Christel Picard, Nicolas Servant, Isabell Rall, Yuvia A. Pérez-Rico, Lena Clerquin, Nila Servaas, Laura Villacorta, Ferris Jung, Christy Luong, Howard Y. Chang, Judith B. Zaugg, Oliver Stegle, Duncan T. Odom, Agnese Loda, Edith Heard
What makes clocks tick? Characterizing developmental dynamics of adult epigenetic clock sites
Rosa H. Mulder, Alexander Neumann, Janine F. Felix, Matthew Suderman, Charlotte A. M. Cecil
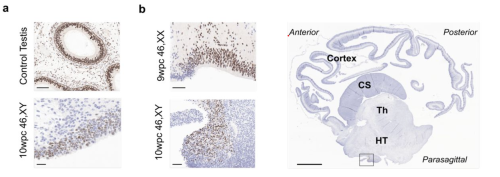
Sex differences in early human fetal brain development
Federica Buonocore, Jenifer P Suntharalingham, Olumide K Ogunbiyi, Aragorn Jones, Nadjeda Moreno, Paola Niola, Tony Brooks, Nita Solanky, Mehul T. Dattani, Ignacio del Valle, John C. Achermann
Tim Casey-Clyde, S. John Liu, Juan Antonio Camara Serrano, Camilla Teng, Yoon-Gu Jang, Harish N. Vasudevan, Jeffrey O. Bush, David R. Raleigh
Yiqiao Zheng, Gary D. Stormo, Shiming Chen
Shruthi Bandyadka, Diane PV Lebo, Albert Mondragon, Sandy B Serizier, Julian Kwan, Jeanne S Peterson, Alexandra Y Chasse, Victoria Jenkins, Anoush Calikyan, Anthony Ortega, Joshua D Campbell, Andrew Emili, Kimberly McCall
Jay N. Joshi, Neha Changela, Lia Mahal, Tyler Defosse, Janet Jang, Lin-Ing Wang, Arunika Das, Joanatta G. Shapiro, Kim McKim
Shaonil Binti, Adison G. Linder, Philip T. Edeen, David S. Fay
Sofia E. Luna, Joab Camarena, Jessica P. Hampton, Kiran R. Majeti, Carsten T. Charlesworth, Eric Soupene, Sridhar Selvaraj, Kun Jia, Vivien A. Sheehan, M. Kyle Cromer, Matthew H. Porteus
Sulzyk Valeria, Curci Ludmila, Lucas N González, Rebagliati Cid Abril, Weigel Muñoz Mariana, Patricia S Cuasnicu
Suvimal Kumar Sindhu, Archita Mishra, Niveda Udaykumar, Jonaki Sen
BCL11b interacts with RNA and proteins involved in RNA processing and developmental diseases
Haitham Sobhy, Marco De Rovere, Amina Ait-Ammar, Muhammad Kashif, Clementine Wallet, Fadoua Daouad, Thomas Loustau, Carine Van Lint, Christian Schwartz, Olivier Rohr
Menghan Wang, Ana Di Pietro-Torres, Christian Feregrino, Maëva Luxey, Chloé Moreau, Sabrina Fischer, Antoine Fages, Patrick Tschopp
Barbara Zhao, Jacob Socha, Andrea Toth, Sharlene Fernandes, Helen Warheit-Niemi, Brandy Ruff, Gurgit K. Khurana Hershey, Kelli L. VanDussen, Daniel Swarr, William J. Zacharias
Hristo Todorov, Stephan Weißbach, Laura Schlichtholz, Hanna Mueller, Dewi Hartwich, Susanne Gerber, Jennifer Winter
| Stem cells, regeneration & disease modelling
Antonia Wiegering, Isabelle Anselme, Ludovica Brunetti, Laura Metayer-Derout, Damelys Calderon, Sophie Thomas, Stéphane Nedelec, Alexis Eschstruth, Sylvie Schneider-Maunoury, Aline Stedman
“Identification of microRNAs regulated by E2F transcription factors in human pluripotent stem cells”
María Soledad Rodríguez-Varela, Mercedes Florencia Vautier, Sofía Mucci, Luciana Isaja, Elmer Fernández, Gustavo Emilio Sevlever, María Elida Scassa, Leonardo Romorini
Fabrication of elastomeric stencils for patterned stem cell differentiation
Stefanie Lehr, Jack Merrin, Monika Kulig, Thomas Minchington, Anna Kicheva
Self-organization of embryonic stem cells into a reproducible embryo model through epigenome editing
Gerrald A. Lodewijk, Sayaka Kozuki, Clara Han, Benjamin R. Topacio, Abolfazl Zargari, Seungho Lee, Gavin Knight, Randolph Ashton, Lei S. Qi, S. Ali Shariati
Transcription factor-based transdifferentiation of human embryonic to trophoblast stem cells
Paula A. Balestrini, Abdelbaki Ahmed, McCarthy Afshan, Liani Devito, Claire E. Senner, Alice E. Chen, Prabhakaran Munusamy, Paul Blakeley, Kay Elder, Phil Snell, Leila Christie, Paul Serhal, Rabi A. Odia, Mahesh Sangrithi, Kathy K. Niakan, Norah M.E. Fogarty
Noura Aldous, Ahmed K. Elsayed, Bushra Memon, Sadaf Ijaz, Sikander Hayat, Essam M. Abdelalim
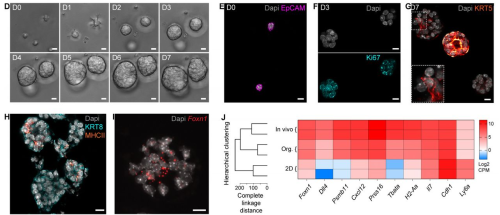
Thymic epithelial organoids mediate T cell development
Tania Hübscher, L. Francisco Lorenzo-Martín, Thomas Barthlott, Lucie Tillard, Jakob J. Langer, Paul Rouse, C. Clare Blackburn, Georg Holländer, Matthias P. Lutolf
Zhiwei Feng, Bingrui Zhou, Qizhi Shuai, Yunliang Wei, Ning Jin, Xiaoling Wang, Hong Zhao, Zhizhen Liu, Jun Xu, Jianbing Mu, Jun Xie
Derivation of elephant induced pluripotent stem cells
Evan Appleton, Kyunghee Hong, Cristina Rodríguez-Caycedo, Yoshiaki Tanaka, Asaf Ashkenazy-Titelman, Ketaki Bhide, Cody Rasmussen-Ivey, Xochitl Ambriz-Peña, Nataly Korover, Hao Bai, Ana Quieroz, Jorgen Nelson, Grishma Rathod, Gregory Knox, Miles Morgan, Nandini Malviya, Kairui Zhang, Brody McNutt, James Kehler, Amanda Kowalczyk, Austin Bow, Bryan McLendon, Brandi Cantarel, Matt James, Christopher E. Mason, Charles Gray, Karl R. Koehler, Virginia Pearson, Ben Lamm, George Church, Eriona Hysolli
Her9 controls the stemness properties of the hindbrain boundary cells
Carolyn Engel-Pizcueta, Covadonga F Hevia, Adrià Voltes, Jean Livet, Cristina Pujades
Regenerative clustering of Enteroblasts in the Drosophila midgut revealed by a morphometric analysis
Fionna Zhu, Michael J. Murray
An Lgr5-independent developmental lineage is involved in mouse intestinal regeneration
Maryam Marefati, Valeria Fernandez-Vallone, Morgane Leprovots, Gabriella Vasile, Frédérick Libert, Anne Lefort, Gilles Dinsart, Achim Weber, Jasna Jetzer, Marie-Isabelle Garcia, Gilbert Vassart
Frédéric Rosa, Nicolas Dray, Laure Bally-Cuif
Rei Yagasaki, Ryo Nakamura, Yuuki Shikaya, Ryosuke Tadokoro, Ruolin Hao, Zhe Wang, Mototsugu Eiraku, Masafumi Inaba, Yoshiko Takahashi
Abdulvasey Mohammed, Benjamin D. Solomon, Priscila F. Slepicka, Kelsea M. Hubka, Hanh Dan Nguyen, Wenqing Wang, Martin Arreola, Michael G. Chavez, Christine Y. Yeh, Doo Kyung Kim, Virginia Winn, Casey A. Gifford, Veronika Kedlian, Jong-Eun Park, Georg A. Hollander, Vittorio Sebastiano, Purvesh Khatri, Sarah A. Teichmann, Andrew J. Gentles, Katja G. Weinacht
Tuft cells act as regenerative stem cells in the human intestine
Lulu Huang, Jochem H. Bernink, Amir Giladi, Daniel Krueger, Gijs J.F. van Son, Maarten H. Geurts, Georg Busslinger, Lin Lin, Maurice Zandvliet, Peter J. Peters, Carmen Lopez-Iglesias, Christianne J. Buskens, Willem A. Bemelman, Harry Begthel, Hans Clevers
Developmental Regulation of Alternative Polyadenylation in an Adult Stem Cell Lineage
Lorenzo Gallicchio, Neuza Reis Matias, Fabian Morales-Polanco, Iliana Nava, Sarah Stern, Yi Zeng, Margaret Theresa Fuller
Human spermatogonial stem cells retain states with a foetal-like signature
Stephen J Bush, Rafail Nikola, Seungmin Han, Shinnosuke Suzuki, Shosei Yoshida, Benjamin D Simons, Anne Goriely
Interplay of IGF1R and estrogen signaling regulates hematopoietic stem and progenitor cells
Ying Xie, Dongxi Xiang, Xin Hu, Hubert Pakula, Eun-Sil Park, Jiadong Chi, Douglas E. Linn, Luwei Tao, Zhe Li
Senolytic CAR T cells reverse aging-associated defects in intestinal regeneration and fitness
Onur Eskiocak, Saria Chowdhury, Vyom Shah, Emmanuella Nnuji-John, Charlie Chung, Jacob A. Boyer, Alexander S. Harris, Jill Habel, Michel Sadelain, Semir Beyaz, Corina Amor
Nuwanthika Wathuliyadde, Katherine E. Willmore, Gregory M. Kelly
Extracellular vesicles promote proliferation in an animal model of regeneration
Priscilla N. Avalos, Lily L. Wong, David J. Forsthoefel
Extended culture of 2D gastruloids to model human mesoderm development
Bohan Chen, Hina Khan, Zhiyuan Yu, LiAng Yao, Emily Freeburne, Kyoung Jo, Craig Johnson, Idse Heemskerk
Sze Hang Kwok, Yuejiang Liu, David Bilder, Jung Kim
Fibroblasts enhance the growth and survival of adult feline small intestinal organoids
Nicole D Hryckowian, Katelyn Studener, Laura J Knoll
Capture of Human Neuromesodermal and Posterior Neural Tube Axial Stem Cells
Dolunay Kelle, Enes Ugur, Ejona Rusha, Dmitry Shaposhnikov, Alessandra Livigni, Sandra Horschitz, Mahnaz Davoudi, Andreas Blutke, Judith Bushe, Michael Sterr, Ksenia Arkhipova, Benjamin Tak, Ruben de Vries, Mazène Hochane, Britte Spruijt, Aicha Haji Ali, Heiko Lickert, Annette Feuchtinger, Philipp Koch, Matthias Mann, Heinrich Leonhardt, Valerie Wilson, Micha Drukker
Marija Lazovska, Kristine Salmina, Dace Pjanova, Bogdan I. Gerashchenko, Jekaterina Erenpreisa
Augusto Ortega Granillo, Daniel Zamora, Robert R. Schnittker, Allison R. Scott, Jonathon Russell, Carolyn E. Brewster, Eric J. Ross, Daniel A. Acheampong, Kevin Ferro, Jason A. Morrison, Boris Y. Rubinstein, Anoja G. Perera, Wei Wang, Alejandro Sánchez Alvarado
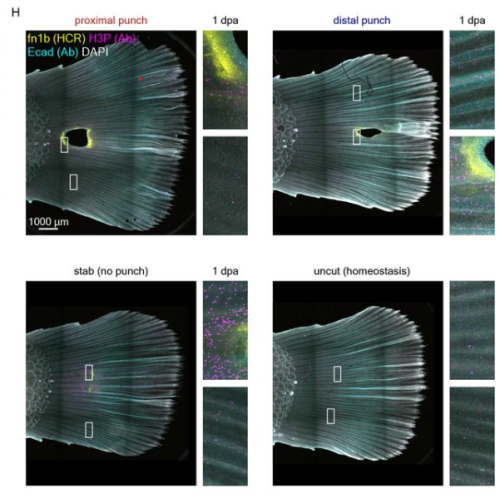
Lucia Leitner, Martina Schultheis, Franziska Hofstetter, Claudia Rudolf, Valeria Kizner, Kerstin Fiedler, Marie-Therese Konrad, Julia Höbaus, Marco Genini, Julia Kober, Elisabeth Ableitner, Teresa Gmaschitz, Diana Walder, Georg Weitzer
Melody Autumn, Yinan Hu, Jenny Zeng, Sarah K. McMenamin
| Plant development
Qian Ma, Sijia Liu, Sara Raggi, Siamsa M. Doyle, Barbora Pařízková, Deepak Kumar Barange, Edward G. Wilkinson, Isidro Crespo Garcia, Joakim Bygdell, Gunnar Wingsle, Dirk Roeland Boer, Lucia C. Strader, Fredrik Almqvist, Ondřej Novák, Stéphanie Robert
Eleanore J. Ritter, Carolyn D. K. Graham, Chad Niederhuth, Marjorie Gail Weber
Xiaosa Xu, Michael Passalacqua, Brian Rice, Edgar Demesa-Arevalo, Mikiko Kojima, Yumiko Takebayashi, Benjamin Harris, Hitoshi Sakakibara, Andrea Gallavotti, Jesse Gillis, David Jackson
Léa Rambaud-Lavigne, Aritra Chatterjee, Simone Bovio, Virginie Battu, Quentin Lavigne, Namrata Gundiah, Arezki Boudaoud, Pradeep Das
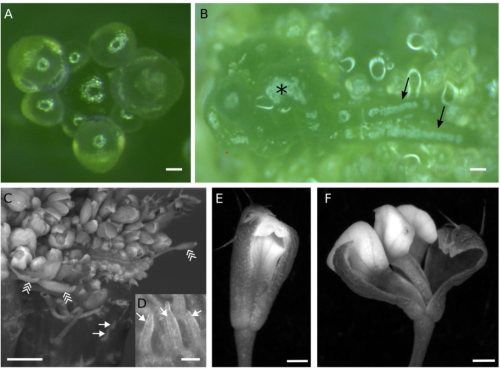
Postembryonic developmental roles of the Arabidopsis KEULE gene
Alejandro Ruiz-Bayón, Carolina Cara-Rodríguez, Raquel Sarmiento-Mañús, Rafael Muñoz-Viana, Francisca M. Lozano, María Rosa Ponce, José Luis Micol
Gerardo del Toro-de León, Joram van Boven, Juan Santos-González, Wen-Biao Jiao, Korbinian Schneeberger, Claudia Köhler
An atlas of Brachypodium distachyon lateral root development
Cristovao De Jesus Vieira Teixeira, Kevin Bellande, Alja van der Schuren, Devin O’Connor, Christian S. Hardtke, Joop EM Vermeer
Uria Ramon, Amit Adiri, Hadar Cheriker, Ido Nir, Yogev Burko, David Weiss
Chiara A. Airoldi, Chao Chen, Humberto Herrera-Ubaldo, Hongbo Fu, Carlos A. Lugo, Alfred J. Crosby, Beverley J. Glover
A RALF-Brassinosteroid morpho-signaling circuit regulates Arabidopsis hypocotyl cell shape
David Biermann, Michelle von Arx, Kaltra Xhelilaj, David Séré, Martin Stegmann, Sebastian Wolf, Cyril Zipfel, Julien Gronnier
| Evo-devo
Rewinding the developmental tape shows how bears break a developmental rule
Otto E. Stenberg, Jacqueline E. Moustakas-Verho, Jukka Jernvall
Avery Leigh Russell, Rosana Zenil-Ferguson, Stephen L. Buchmann, Diana D. Jolles, Ricardo Kriebel, Mario Vallejo-Marín
Comparative analysis of Wolbachia maternal transmission and localization in host ovaries
Michael T.J. Hague, Timothy B. Wheeler, Brandon S. Cooper
Paula R. Roy, Dean M. Castillo
A male-essential microRNA is key for avian sex chromosome dosage compensation
Amir Fallahshahroudi, Leticia Rodríguez-Montes, Sara Yousefi Taemeh, Nils Trost, Memo Tellez Jr., Maeve Ballantyne, Alewo Idoko-Akoh, Lorna Taylor, Adrian Sherman, Enrico Sorato, Martin Johnsson, Margarida Cardoso Moreira, Mike J. McGrew, Henrik Kaessmann
Repatterning of mammalian backbone regionalization in cetaceans
Amandine Gillet, Katrina E. Jones, Stephanie E. Pierce
A multicellular developmental program in a close animal relative
Marine Olivetta, Chandni Bhickta, Nicolas Chiaruttini, John Burns, Omaya Dudin
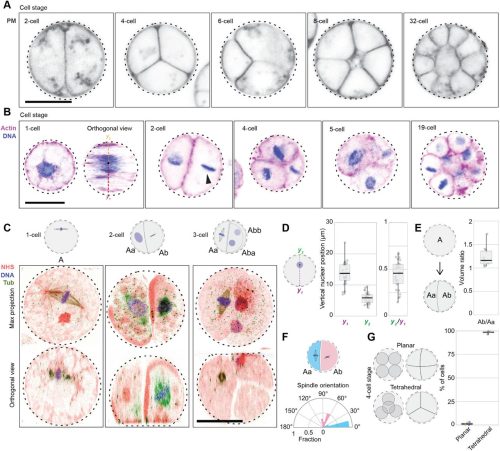
Gene regulatory network co-option is sufficient to induce a morphological novelty in Drosophila
Gavin Rice, Tatiana Gaitan-Escudero, Kenechukwu Charles-Obi, Julia Zeitlinger, Mark Rebeiz
Aurélie Hintermann, Christopher Chase Bolt, M. Brent Hawkins, Guillaume Valentin, Lucille Lopez-Delisle, Sandra Gitto, Paula Barrera Gómez, Bénédicte Mascrez, Thomas A. Mansour, Tetsuya Nakamura, Matthew P. Harris, Neil H. Shubin, Denis Duboule
Independent size regulation of bones and appendages in zebrafish
Toshihiro Aramaki, Shigeru Kondo
Cell Biology
Neutral evolution of snoRNA Host Gene long non-coding RNA affects cell fate control
Matteo Vietri Rudan, Kalle H. Sipilä, Christina Philippeos, Clarisse Gânier, Victor A. Negri, Fiona M. Watt
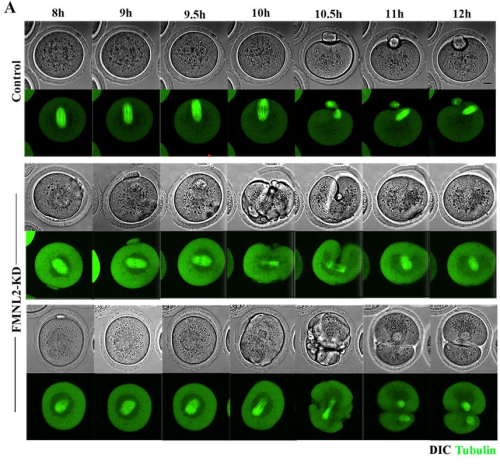
FMNL2 regulates actin for ER and mitochondria distribution in oocyte meiosis
Meng-Hao Pan, Zhen-Nan Pan, Ming-Hong Sun, Xiao-Han Li, Jia-Qian Ju, Shi-Ming Luo, Xiang-Hong Ou, Shao-Chen Sun
Aurora B controls microtubule stability to regulate abscission dynamics in stem cells
Snježana Kodba, Amber Öztop, Eri van Berkum, Malina K. Iwanski, Wilco Nijenhuis, Lukas C. Kapitein, Agathe Chaigne
Mitochondrial dynamics regulate cell size in the developing cochlea
James D. B. O’Sullivan, Stephen Terry, Claire A. Scott, Anwen Bullen, Daniel J. Jagger, Zoë F. Mann
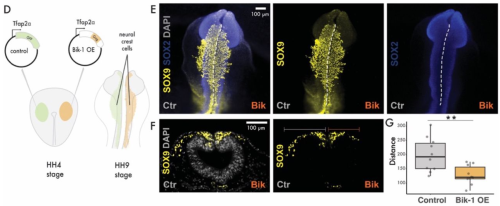
Cell extrusion – a novel mechanism driving neural crest cell delamination
Emma Moore, Ruonan Zhao, Mary C McKinney, Kexi Yi, Christopher Wood, Paul Trainor
Florian Constanty, Bailin Wu, Ke-Hsuan Wei, I-Ting Lin, Julia Dallmann, Stefan Guenther, Till Lautenschlaeger, Rashmi Priya, Shih-Lei Lai, Didier Y.R. Stainier, Arica Beisaw
Yi-Ting Huang, Lauren L. Hesting, Brian R. Calvi
Steven L. Brody, Jiehong Pan, Tao Huang, Jian Xu, Huihui Xu, Jeffrey Koenitizer, Steven K. Brennan, Rashmi Nanjundappa, Thomas G. Saba, Andrew Berical, Finn J. Hawkins, Xiangli Wang, Rui Zhang, Moe R. Mahjoub, Amjad Horani, Susan K. Dutcher
Pooja Popli, Arin K. Oestreich, Vineet K. Maurya, Marina N. Rowen, Ramya Masand, Michael J. Holtzman, Yong Zhang, John Lydon, Shizuo Akira, Kelle H. Moley, Ramakrishna Kommagani
Defining the contribution of Troy-positive progenitor cells to the mouse esophageal epithelium
David Grommisch, Menghan Wang, Evelien Eenjes, Maja Svetličič, Qiaolin Deng, Pontus Giselsson, Maria Genander
Shruthi Bandyadka, Diane PV Lebo, Albert Mondragon, Sandy B Serizier, Julian Kwan, Jeanne S Peterson, Alexandra Y Chasse, Victoria Jenkins, Anoush Calikyan, Anthony Ortega, Joshua D Campbell, Andrew Emili, Kimberly McCall
Gag proteins encoded by endogenous retroviruses are required for zebrafish development
Ni-Chen Chang, Jonathan N. Wells, Andrew Y. Wang, Phillip Schofield, Yi-Chia Huang, Vinh H. Truong, Marcos Simoes-Costa, Cédric Feschotte
FilamentID reveals the composition and function of metabolic enzyme polymers during gametogenesis
Jannik Hugener, Jingwei Xu, Rahel Wettstein, Lydia Ioannidi, Daniel Velikov, Florian Wollweber, Adrian Henggeler, Joao Matos, Martin Pilhofer
Desynchronization between timers provokes transient arrest during C. elegans development
Francisco J. Romero-Expósito, Almudena Moreno-Rivero, Marta Muñoz-Barrera, Sabas García-Sánchez, Fernando Rodríguez-Peris, Nicola Gritti, Francesca Sartor, Martha Merrow, Jeroen S. van Zon, Alejandro Mata-Cabana, María Olmedo
Modelling
Manica Balant, Teresa Garnatje, Daniel Vitales, Oriane Hidalgo, Daniel H. Chitwood
Yusuke Sakai, Jun Hakura
Pamela C. Guruciaga, Takafumi Ichikawa, Takashi Hiiragi, Anna Erzberger
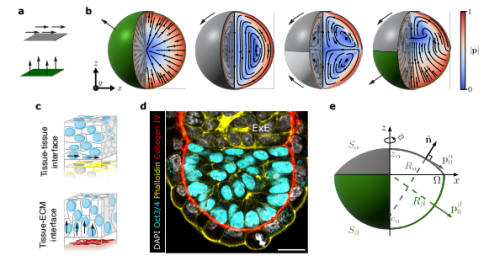
Mechanochemical bistability of intestinal organoids enables robust morphogenesis
Shi-Lei Xue, Qiutan Yang, Prisca Liberali, Edouard Hannezo
Waves, patterns and bifurcations: a tutorial review on the vertebrate segmentation clock
Paul François, Victoria Mochulska
Michael A. Ramirez-Sierra, Thomas R. Sokolowski
Tools & Resources
Deep learning methods to forecasting human embryo development in time-lapse videos
Akriti Sharma, Alexandru Dorobantiu, Saquib Ali, Mario Iliceto, Mette H. Stensen, Erwan Delbarre, Michael A. Riegler, Hugo L. Hammer
Comprehensive mapping of sensory and sympathetic innervation of the developing kidney
Pierre-Emmanuel Y. N’Guetta, Sarah R. McLarnon, Adrien Tassou, Matan Geron, Sepenta Shirvan, Rose Z. Hill, Grégory Scherrer, Lori L. O’Brien
EyeHex toolbox for complete segmentation of ommatidia in fruit fly eyes
Huy Tran, Nathalie Dostatni, Ariane Ramaekers
Spatial Dynamics of the Developing Human Heart
Enikő Lázár, Raphaël Mauron, Žaneta Andrusivová, Julia Foyer, Ludvig Larsson, Nick Shakari, Sergio Marco Salas, Sanem Sariyar, Jan N. Hansen, Marco Vicari, Paulo Czarnewski, Emelie Braun, Xiaofei Li, Olaf Bergmann, Christer Sylvén, Emma Lundberg, Sten Linnarsson, Mats Nilsson, Erik Sundström, Igor Adameyko, Joakim Lundeberg
Transcriptional dynamics of the murine heart during perinatal development at single-cell resolution
Lara Feulner, Florian Wünnemann, Jenna Liang, Philipp Hofmann, Marc-Phillip Hitz, Denis Schapiro, Severine Leclerc, Patrick Piet van Vliet, Gregor Andelfinger
Xianfeng Yang, Qiufei Lin, Jinu Udayabhanu, Yuwei Hua, Xuemei Dai, Shichao Xin, Huasun Huang, Tiandai Huang
D. Nathaniel Clarke, Laurent Formery, Christopher J Lowe
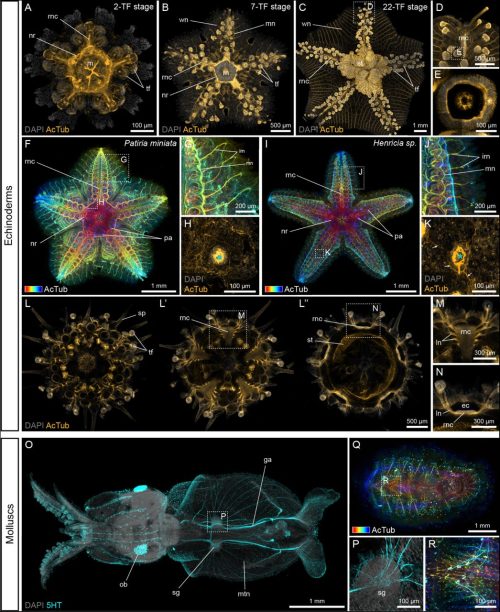
Ke Zhang, Yanqiu Wang, Qi An, Hengjing Ji, Defu Wu, Xuri Li, Xuran Dong, Chun Zhang
Highly efficient tamoxifen-inducible Cre recombination in embryonic, larval and adult zebrafish
Edita Bakūnaitė, Emilija Gečaitė, Justas Lazutka, Darius Balciunas
Camilo V. Echeverria Jr., Tess A. Leathers, Crystal D. Rogers
Elliot W. Jackson, Emilio Romero, Svenja Kling, Yoon Lee, Evan Tjeerdema, Amro Hamdoun
A high-throughput method for quantifying Drosophila fecundity
Andreana Gomez, Sergio Gonzalez, Ashwini Oke, Jiayu Luo, Johnny B. Duong, Raymond M. Esquerra, Thomas Zimmerman, Sara Capponi, Jennifer C. Fung, Todd G. Nystul
Research practice & education
CRISPR/Cas9 gene editing in Drosophila via visual selection in a summer classroom
Lutz Kockel, Valentina Zhang, Jenna Wang, Clara Gulick, Madeleine E. Laws, Arjun Rajan, Nicole Lantz, Ayla Asgarova, Lillian Dai, Kristian Garcia, Charlene Kim, Michelle Li, Patricio Ordonez-Acosta, Dongshen Peng, Henry Shull, Lauren Tse, Yixang Wang, Wenxin Yu, Zee Zhou, Anne Rankin, Sangbin Park, Seung K. Kim
Australian researchers’ perceptions and experiences with stem cell registration
Mengqi Hu, Dan Santos, Edilene Lopes, Dianne Nicol, Andreas Kurtz, Nancy Mah, Sabine C. Muller, Rachel A. Ankeny, Christine A. Wells
Katy Andrews, Rosalie Stoneley, Katja Eckl
Rendering protein structures inside cells at the atomic level with Unreal Engine
Muyuan Chen
Perry G. Beasley-Hall, Pam Papadelos, Anne Hewitt, Charlotte R. Lassaline, Kate D. L. Umbers, Michelle T. Guzik


 (No Ratings Yet)
(No Ratings Yet)



 (1 votes)
(1 votes)