Hi I’m Steve and I work in the Sensory Development lab at the RIKEN Center for Developmental Biology in Kobe, Japan. We study the development of the chick inner ear. I haven’t always been in a chick lab – during my Ph.D. at The University of York I used Xenopus, so it was really nice to read Gary’s post. Looking back, I miss those slimy yet serene critters. Although at the time (as a poor PhD student) I would often get pretty jealous of all the care, attention, and, in particular, premium food they used to get. Especially when I was tucking into my staple Ph.D. meal, the supernoodle sandwich™. Anyway, enough of the reminiscing!
Chick-en you believe it?
The chick is a brilliant and beautiful model system for studying the embryonic development, and in our case more specifically, the inner ear. Embryo husbandry could not be simpler, because the hen is thoughtful enough to pack everything the embryo needs to develop inside the egg. We simply buy fertilized eggs from a local farm, incubate them at 37°C, and hey presto, the embryos develop!

It takes 21 days for chicks to hatch, but luckily we don’t have to wait that long if we need chicks, because we can also order 20 day-old fertilized eggs that arrive ready to hatch. When hatching, the chick cuts through the top of the shell using its “egg tooth” and then shimmies out. This takes a lot of work, so the chicks are pretty exhausted afterwards and they often take a little nap. But once they recover they are soon up and at ‘em, hopping around and pecking for food.
Our hatched chicks reside in a special bird room where they live in large temperature controlled incubators complete with sawdust and shredded paper bedding, fresh clean water, and mix of cereal and seed for food. They get fresh bedding, food and water every day, and we are lucky to have a very attentive technician (she calls herself “Mama Hen”) who takes care great of them.

Access all areas.
Because chick embryos develop ex-utero, we have easy access to them from the very earliest stages of development. We put a small strip of sellotape along the shell, cut a window through it, and can access the embryo through this window as it develops on the surface of the yolk. This unlimited access is fantastic for us, because the inner ear is specified very early in development. To understand how these events occur, we need to be able to manipulate the system at these early stages. The chick system makes it easy to do so.
Spot the difference.
Mice are the kings when it comes to using a model system close to humans. So why bother using other systems at all? Well, one reason is that the differences between species can be just as important as the similarities. Birds, unlike mammals, can regenerate the hair cells of their inner ear, so for our lab this makes them a great system to use for researching the mechanisms that govern regeneration. We use hatched chicks up to around 3 weeks old to study regeneration. By focusing on the differences as well as they similarities between species we can gain an insight into what exactly are the key mechanisms that coordinate the regenerative process.
An egg-sample day – the morning.
Ok, no more puns, I promise. The first thing I do when I arrive in the morning is check the embryos already incubating from previous day’s experiments. It is important to keep the incubator as clean as possible, so we keep a keen eye out for any signs of infection and remove the offending eggs.
Next I will check to see if any fresh fertilized eggs have arrived for me. If so, I put them in a 14°C fridge. This arrests the embryos’ development, and allows me to have a better idea of their developmental time course when I come to incubate them at 37°C. Accurate timing is everything because we don’t have near limitless supplies of eggs. We receive egg deliveries twice a week, and you have to order your eggs a week in advance, so forward planning is critical. If you miss the stage you want, you have to wait until next weeks eggs arrive to start again.
After this, I usually start some electroporation experiments. I like to do these in the morning, for two reasons. Firstly, I am usually electroporating something that is GFP or RFP tagged, so a morning session means I can check for fluorescence just before I go home in the evening. Secondly, the ever-approaching lunchtime serves as good motivation to work quickly.
Our electroporation set up is in the main area of our lab, near to a gargantuan egg incubator and a darkroom for fluorescent microscopy. A common and somewhat surreal result of this set up is that I often find myself chatting with someone who has their head (and often most of their upper body) inside the incubator as they check their eggs. To increase the surreal stakes further, what appears to be a dismembered head will often join the conversation when someone who is working in the dark room sticks their head around the curtain to get involved. And of course I’m trying to talk with a mouth pipette for the electroporations in my mouth. Despite (or maybe because of) this, it is one of my favourite areas of the lab.
The afternoon
The afternoons are spent doing a variety of things, depending on what stage of the project I am at and what the priorities are. Often I am finishing up ongoing experiments, – doing some immunohistochemistry, some confocal microscopy, analyzing data, administering drug treatments, fixing samples, or doing some in situ hybridisations. Actually, on the subject of in situ’s, we have an “in situ robot”, which runs through most of the protocol autonomously, leaving us with the job of overseeing the final colour reaction. I’m pretty skeptical about this robot – something about which the rest of the lab enjoy mercilessly taking the mickey out of me. But I’ve seen the Terminator movies, and I know what happens when robots get too smart. This one hasn’t done anything other than produce beautiful gene expression patterns yet, but it is only a matter of time.
The day ends incubating some more eggs for the next set of experiments. This is probably the most dangerous time of the day for me. The fridge and the incubator are at opposite ends of the lab, so I run a nightly gauntlet from one to the other, precariously tippy toeing through the busy lab with trays of eggs in my hands. Each tray carries 24 eggs, and when you drop one (as I have many times over the years) it really sucks!
 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series here and read other posts in this series here.
 (13 votes)
(13 votes)
 Loading...
Loading...


 (3 votes)
(3 votes)

 This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series
This post is part of a series on a day in the life of developmental biology labs working on different model organisms. You can read the introduction to the series 

 (No Ratings Yet)
(No Ratings Yet)
 Mammalian cardiac regeneration is greatly impeded by the massive loss of cardiomyocytes that occurs following acute injury. The failure of the remaining cells to proliferate is a considerable challenge for the field, but the molecular mechanisms that control cardiomyocyte proliferation in the adult heart are largely unknown. Now, on
Mammalian cardiac regeneration is greatly impeded by the massive loss of cardiomyocytes that occurs following acute injury. The failure of the remaining cells to proliferate is a considerable challenge for the field, but the molecular mechanisms that control cardiomyocyte proliferation in the adult heart are largely unknown. Now, on 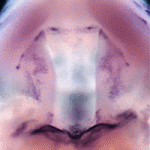 Precise orchestration of palate formation involves the complex interaction of signalling cascades and transcriptional networks in the developing craniofacial region. Pax9 and Osr2 have previously been implicated in palate formation, but little is known about how these molecular components interact within the greater regulatory network. Now, on
Precise orchestration of palate formation involves the complex interaction of signalling cascades and transcriptional networks in the developing craniofacial region. Pax9 and Osr2 have previously been implicated in palate formation, but little is known about how these molecular components interact within the greater regulatory network. Now, on 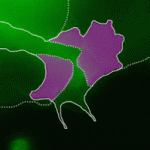 Contact inhibition of locomotion (CIL) is a fundamental regulatory mechanism that ensures correct cell movement and migration. During CIL, cells form transient contacts but the molecular nature of such contacts is unknown. In this issue, Roberto Mayor and colleagues (
Contact inhibition of locomotion (CIL) is a fundamental regulatory mechanism that ensures correct cell movement and migration. During CIL, cells form transient contacts but the molecular nature of such contacts is unknown. In this issue, Roberto Mayor and colleagues (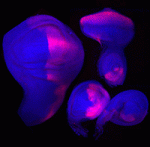 The immense power of Drosophila genetics has allowed invaluable insight into developmental biology. Despite these advances, a significant limitation has always been the lack of an efficient method for modifying select genetic loci. Now, on
The immense power of Drosophila genetics has allowed invaluable insight into developmental biology. Despite these advances, a significant limitation has always been the lack of an efficient method for modifying select genetic loci. Now, on 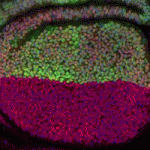 Proper control of cell size is vital to ensure the correct growth and development of any organism. The Myc family of proteins are key regulators of growth, but the mechanisms that control Myc protein levels are complex. Now, on
Proper control of cell size is vital to ensure the correct growth and development of any organism. The Myc family of proteins are key regulators of growth, but the mechanisms that control Myc protein levels are complex. Now, on 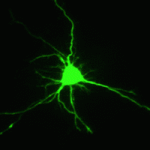 Dendrite complexity determines the functional properties of neurons and the overall connectivity of neuronal circuits. The bone morphogenetic protein (BMP) family is known to regulate a myriad of developmental processes, but the extent to which different members of the family are involved in dendrite growth remains unclear. In this issue (
Dendrite complexity determines the functional properties of neurons and the overall connectivity of neuronal circuits. The bone morphogenetic protein (BMP) family is known to regulate a myriad of developmental processes, but the extent to which different members of the family are involved in dendrite growth remains unclear. In this issue ( Stem cells and their progenitors are maintained within a microenvironment, termed the niche, but it is known that systemic signals originating outside the niche also affect stem cell and progenitor behavior. Here, Utpal Banerjee and colleagues review recent studies of nutritional effects on stem and progenitor cell maintenance and proliferation in Drosophila. See the Review article on p.
Stem cells and their progenitors are maintained within a microenvironment, termed the niche, but it is known that systemic signals originating outside the niche also affect stem cell and progenitor behavior. Here, Utpal Banerjee and colleagues review recent studies of nutritional effects on stem and progenitor cell maintenance and proliferation in Drosophila. See the Review article on p.  The proper formation and morphogenesis of dendrites is fundamental to the establishment of neural circuits in the brain. In this issue, Sidharth Puram and Azad Bonni review cell-intrinsic drivers of dendrite patterning and discuss how the characterization of such regulators advances our understanding of normal brain development and pathogenesis of diverse cognitive disorders. See the Review on p.
The proper formation and morphogenesis of dendrites is fundamental to the establishment of neural circuits in the brain. In this issue, Sidharth Puram and Azad Bonni review cell-intrinsic drivers of dendrite patterning and discuss how the characterization of such regulators advances our understanding of normal brain development and pathogenesis of diverse cognitive disorders. See the Review on p.