Here are the highlights from the current issue of Development:
Haematopoiesis to knock your SOX7 off
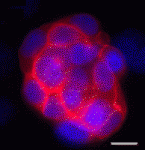
During vertebrate development, haematopoietic and endothelial cells emerge from a common mesodermal progenitor, the haemangioblast. Upon haematopoietic commitment, the haemangioblast generates blood precursors through populations of endothelial cells with haemogenic properties, but what regulates the generation of haemogenic endothelium? Here (
p. 1587), Valerie Kouskoff and colleagues use mouse embryonic stem cells to investigate the role of the transcription factor SOX7 in haemangioblast differentiation. The researchers report that, within blast colonies (which recapitulate early haematopoietic development in vitro), SOX7 is expressed in haemogenic endothelium cells and is downregulated in nascent blood precursors. Enforced expression of
Sox7 in blast colonies, they report, blocks haematopoietic differentiation and sustains the expression of endothelial markers. They also show that SOX7 binds and activates the promoter of VE-cadherin, an adhesion molecule that is expressed in haemogenic endothelial cells but downregulated during blood cell formation. Together, these results identify SOX7 as a transcriptional regulator of genes expressed in the haemogenic endothelium and provide new insights into early embryonic haematopoiesis.
A novel photo-morpholino on/off switch for genes
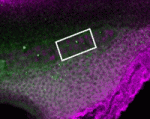 To investigate the molecular mechanisms of development, it is useful to be able to turn genes both on and off in a spatially restricted manner. Although the development of photo-activated morpholino oligonucleotides (photo-MOs) has made it possible to turn genes off at specific times and places, finding a way to deactivate MOs and restart gene expression has proved more elusive. Here (p.1691), Alexandra Tallafuss, Philip Washbourne and colleagues describe an approach in which they can turn genes off and on using sense photo-MOs (S-photo-MOs) and complementary antisense photo-MOs (AS-photo-MOs), respectively. S-photo-MOs bind to and block the activity of regular morpholinos, and exposure of the S-photo-MOs to UV light allows the morpholinos to become active and block gene expression; AS-photo-MOs block gene function like regular morpholinos, but normal gene function can be restored by light inactivation. Importantly, the researchers demonstrate the feasibility of their new approach in whole zebrafish embryos by studying notochord induction and neural crest development, and in single cells by temporally manipulating Gal4 transgene expression.
To investigate the molecular mechanisms of development, it is useful to be able to turn genes both on and off in a spatially restricted manner. Although the development of photo-activated morpholino oligonucleotides (photo-MOs) has made it possible to turn genes off at specific times and places, finding a way to deactivate MOs and restart gene expression has proved more elusive. Here (p.1691), Alexandra Tallafuss, Philip Washbourne and colleagues describe an approach in which they can turn genes off and on using sense photo-MOs (S-photo-MOs) and complementary antisense photo-MOs (AS-photo-MOs), respectively. S-photo-MOs bind to and block the activity of regular morpholinos, and exposure of the S-photo-MOs to UV light allows the morpholinos to become active and block gene expression; AS-photo-MOs block gene function like regular morpholinos, but normal gene function can be restored by light inactivation. Importantly, the researchers demonstrate the feasibility of their new approach in whole zebrafish embryos by studying notochord induction and neural crest development, and in single cells by temporally manipulating Gal4 transgene expression.
See also the authors’ post about this paper on the Node.
Endoderm tugs at heart strings
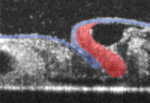 During vertebrate heart development, cardiac progenitors form bilateral fields within the lateral plate mesoderm that move towards the embryonic midline and fuse to form the heart tube. It is generally accepted that, during this process, in addition to its inductive signalling role, the endoderm serves as a mechanically passive substrate for the migrating cardiogenic mesoderm. Now, Victor Varner and Larry Taber suggest that the endoderm also has an active mechanical role during heart tube assembly (see p. 1680). Using label-tracking experiments in combination with actomyosin inhibitors, the researchers show that, in chick embryos, the endoderm actively shortens around the anterior intestinal portal, and that myosin-II-dependent contraction drives this shortening. Inhibition of contractility using actomyosin inhibitors, they report, produces cardia bifida and foregut defects. Moreover, computational modelling suggests that the endoderm (not the mesoderm) is the dominant contractile tissue layer during heart field migration. Thus, the researchers suggest, during avian cardiogenesis, active contraction of the endoderm pulls the heart fields towards the embryonic midline.
During vertebrate heart development, cardiac progenitors form bilateral fields within the lateral plate mesoderm that move towards the embryonic midline and fuse to form the heart tube. It is generally accepted that, during this process, in addition to its inductive signalling role, the endoderm serves as a mechanically passive substrate for the migrating cardiogenic mesoderm. Now, Victor Varner and Larry Taber suggest that the endoderm also has an active mechanical role during heart tube assembly (see p. 1680). Using label-tracking experiments in combination with actomyosin inhibitors, the researchers show that, in chick embryos, the endoderm actively shortens around the anterior intestinal portal, and that myosin-II-dependent contraction drives this shortening. Inhibition of contractility using actomyosin inhibitors, they report, produces cardia bifida and foregut defects. Moreover, computational modelling suggests that the endoderm (not the mesoderm) is the dominant contractile tissue layer during heart field migration. Thus, the researchers suggest, during avian cardiogenesis, active contraction of the endoderm pulls the heart fields towards the embryonic midline.
Notch tunes pancreatic endocrine progenitor behaviour
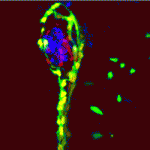 During pancreatic development in mammals and zebrafish, Notch-responsive cells (NRCs) give rise to endocrine cells, but how does Notch signalling regulate the proliferation and differentiation of pancreatic progenitors at the single-cell level? On p. 1557, Nikolay Ninov and co-workers use live imaging and a novel transgenic reporter system that allows the dynamic assessment of Notch signalling to answer this question in zebrafish larvae. The researchers show that zebrafish NRCs exhibit different levels of Notch signalling that, in turn, regulate distinct cellular outcomes. Specifically, high levels of Notch signalling induce quiescence, lower levels promote progenitor amplification, and sustained downregulation of Notch signalling triggers a multistep process that includes cell-cycle entry and amplification followed by endocrine differentiation. Importantly, the researchers show that progenitor amplification and differentiation can be uncoupled by modulating the duration and/or level of Notch signalling downregulation. Thus, different levels of Notch signalling drive distinct behaviours in pancreatic endocrine precursors, a finding that could inform the development of regenerative therapies for type 1 diabetes.
During pancreatic development in mammals and zebrafish, Notch-responsive cells (NRCs) give rise to endocrine cells, but how does Notch signalling regulate the proliferation and differentiation of pancreatic progenitors at the single-cell level? On p. 1557, Nikolay Ninov and co-workers use live imaging and a novel transgenic reporter system that allows the dynamic assessment of Notch signalling to answer this question in zebrafish larvae. The researchers show that zebrafish NRCs exhibit different levels of Notch signalling that, in turn, regulate distinct cellular outcomes. Specifically, high levels of Notch signalling induce quiescence, lower levels promote progenitor amplification, and sustained downregulation of Notch signalling triggers a multistep process that includes cell-cycle entry and amplification followed by endocrine differentiation. Importantly, the researchers show that progenitor amplification and differentiation can be uncoupled by modulating the duration and/or level of Notch signalling downregulation. Thus, different levels of Notch signalling drive distinct behaviours in pancreatic endocrine precursors, a finding that could inform the development of regenerative therapies for type 1 diabetes.
Apical domain size matters for neurogenesis
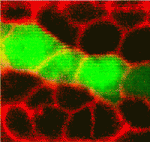 Early in retinal development, neuroepithelial progenitor cells begin to divide in a neurogenic mode in which at least one daughter cell exits the cell cycle and differentiates into a neuron. Now, on p. 1599, Brian Link and colleagues investigate the cell biological mechanisms that influence neurogenesis by analysing zebrafish and mouse retinal neuroepithelia deficient for Llgl1 (lethal giant larval protein homologue 1), a protein that is implicated in apicobasal polarity, asymmetric division, cell shape and cell-cycle exit. The researchers show that Llgl1-deficient neuroepithelia retain apicobasal polarity but have expanded apical domains. Importantly, Llgl1-deficient neuroepithelia display increased Notch activity and reduced neurogenesis. Expansion of the apical domain through inhibition of Shroom3 also increases Notch activity and reduces neurogenesis. These results, together with studies of interkinetic nuclear migration (the apical-to-basal movement of neuroepithelial nuclei that is linked with cell-cycle exit) in Llgl1-depleted retinal precursors, suggest that the size of the apical domain of retinal neuroepithelia modulates the strength of the polarised signals that influence neurogenesis.
Early in retinal development, neuroepithelial progenitor cells begin to divide in a neurogenic mode in which at least one daughter cell exits the cell cycle and differentiates into a neuron. Now, on p. 1599, Brian Link and colleagues investigate the cell biological mechanisms that influence neurogenesis by analysing zebrafish and mouse retinal neuroepithelia deficient for Llgl1 (lethal giant larval protein homologue 1), a protein that is implicated in apicobasal polarity, asymmetric division, cell shape and cell-cycle exit. The researchers show that Llgl1-deficient neuroepithelia retain apicobasal polarity but have expanded apical domains. Importantly, Llgl1-deficient neuroepithelia display increased Notch activity and reduced neurogenesis. Expansion of the apical domain through inhibition of Shroom3 also increases Notch activity and reduces neurogenesis. These results, together with studies of interkinetic nuclear migration (the apical-to-basal movement of neuroepithelial nuclei that is linked with cell-cycle exit) in Llgl1-depleted retinal precursors, suggest that the size of the apical domain of retinal neuroepithelia modulates the strength of the polarised signals that influence neurogenesis.
Mitotic-meiotic switch awry in testicular teratoma
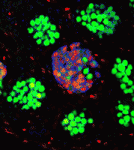 The formation of testicular teratomas and other testicular germ cell tumours (TGCTs) is thought to involve defects in male germ cell development. On p. 1577, Jason Heaney and co-workers provide new insights into TGCT development by studying strains of mice with different teratoma incidences, including the 129 strain, which has a high spontaneous rate of TGCTs. In 129 mice, TGCTs are visible at embryonic day (E) 15.5 as microscopic foci of pluripotent stem cells (embryonal carcinoma cells). The researchers report that germ cell proliferation, the expression of the pluripotency factor Nanog and the premature expression of genes normally expressed in pre-meiotic adult male germ cells, such as cyclin D1 and Stra8, at E15.5 are all directly related to increased tumour risk. Notably, embryonal carcinoma cells co-express genes involved in germ cell pluripotency and differentiation, and deletion of Stra8 in 129 mice reduces their teratoma incidence. Together, these results indicate that deregulation of the mitotic-meiotic switch in male germ cells contributes to teratoma initiation.
The formation of testicular teratomas and other testicular germ cell tumours (TGCTs) is thought to involve defects in male germ cell development. On p. 1577, Jason Heaney and co-workers provide new insights into TGCT development by studying strains of mice with different teratoma incidences, including the 129 strain, which has a high spontaneous rate of TGCTs. In 129 mice, TGCTs are visible at embryonic day (E) 15.5 as microscopic foci of pluripotent stem cells (embryonal carcinoma cells). The researchers report that germ cell proliferation, the expression of the pluripotency factor Nanog and the premature expression of genes normally expressed in pre-meiotic adult male germ cells, such as cyclin D1 and Stra8, at E15.5 are all directly related to increased tumour risk. Notably, embryonal carcinoma cells co-express genes involved in germ cell pluripotency and differentiation, and deletion of Stra8 in 129 mice reduces their teratoma incidence. Together, these results indicate that deregulation of the mitotic-meiotic switch in male germ cells contributes to teratoma initiation.
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...
 A postdoctoral position studying skeletal development is available in the laboratory of Dr. Amy Merrill at the University of Southern California’s Center for Craniofacial Molecular Biology (CCMB). CCMB offers a highly supportive and interactive environment with a strong research profile in craniofacial development and repair. Work conducted in the Merrill laboratory integrates human genetics and developmental biology to study normal and abnormal craniofacial development. We are looking for an enthusiastic candidate who will carry out an independent research project aimed at understanding spatiotemporal signals that pattern bone and cartilage. Ideal applicants will be highly motivated and have recently completed doctoral training in Developmental Biology.
A postdoctoral position studying skeletal development is available in the laboratory of Dr. Amy Merrill at the University of Southern California’s Center for Craniofacial Molecular Biology (CCMB). CCMB offers a highly supportive and interactive environment with a strong research profile in craniofacial development and repair. Work conducted in the Merrill laboratory integrates human genetics and developmental biology to study normal and abnormal craniofacial development. We are looking for an enthusiastic candidate who will carry out an independent research project aimed at understanding spatiotemporal signals that pattern bone and cartilage. Ideal applicants will be highly motivated and have recently completed doctoral training in Developmental Biology. 

 (1 votes)
(1 votes)

 (No Ratings Yet)
(No Ratings Yet)


 During vertebrate development, haematopoietic and endothelial cells emerge from a common mesodermal progenitor, the haemangioblast. Upon haematopoietic commitment, the haemangioblast generates blood precursors through populations of endothelial cells with haemogenic properties, but what regulates the generation of haemogenic endothelium? Here (
During vertebrate development, haematopoietic and endothelial cells emerge from a common mesodermal progenitor, the haemangioblast. Upon haematopoietic commitment, the haemangioblast generates blood precursors through populations of endothelial cells with haemogenic properties, but what regulates the generation of haemogenic endothelium? Here ( To investigate the molecular mechanisms of development, it is useful to be able to turn genes both on and off in a spatially restricted manner. Although the development of photo-activated morpholino oligonucleotides (photo-MOs) has made it possible to turn genes off at specific times and places, finding a way to deactivate MOs and restart gene expression has proved more elusive. Here (
To investigate the molecular mechanisms of development, it is useful to be able to turn genes both on and off in a spatially restricted manner. Although the development of photo-activated morpholino oligonucleotides (photo-MOs) has made it possible to turn genes off at specific times and places, finding a way to deactivate MOs and restart gene expression has proved more elusive. Here ( During vertebrate heart development, cardiac progenitors form bilateral fields within the lateral plate mesoderm that move towards the embryonic midline and fuse to form the heart tube. It is generally accepted that, during this process, in addition to its inductive signalling role, the endoderm serves as a mechanically passive substrate for the migrating cardiogenic mesoderm. Now, Victor Varner and Larry Taber suggest that the endoderm also has an active mechanical role during heart tube assembly (see
During vertebrate heart development, cardiac progenitors form bilateral fields within the lateral plate mesoderm that move towards the embryonic midline and fuse to form the heart tube. It is generally accepted that, during this process, in addition to its inductive signalling role, the endoderm serves as a mechanically passive substrate for the migrating cardiogenic mesoderm. Now, Victor Varner and Larry Taber suggest that the endoderm also has an active mechanical role during heart tube assembly (see  During pancreatic development in mammals and zebrafish, Notch-responsive cells (NRCs) give rise to endocrine cells, but how does Notch signalling regulate the proliferation and differentiation of pancreatic progenitors at the single-cell level? On
During pancreatic development in mammals and zebrafish, Notch-responsive cells (NRCs) give rise to endocrine cells, but how does Notch signalling regulate the proliferation and differentiation of pancreatic progenitors at the single-cell level? On  Early in retinal development, neuroepithelial progenitor cells begin to divide in a neurogenic mode in which at least one daughter cell exits the cell cycle and differentiates into a neuron. Now, on
Early in retinal development, neuroepithelial progenitor cells begin to divide in a neurogenic mode in which at least one daughter cell exits the cell cycle and differentiates into a neuron. Now, on  The formation of testicular teratomas and other testicular germ cell tumours (TGCTs) is thought to involve defects in male germ cell development. On
The formation of testicular teratomas and other testicular germ cell tumours (TGCTs) is thought to involve defects in male germ cell development. On