Gene expansions underlying placenta evolution
Posted by Prahalad Giridhar, on 12 December 2022
As part of the Crick-Calleva program, I had the opportunity to work with Greg Slodkowicz in Margarida Cardoso-Moreira’s lab at the Francis Crick Institute over the past summer, studying the molecular mechanisms underpinning the evolution of the placenta.
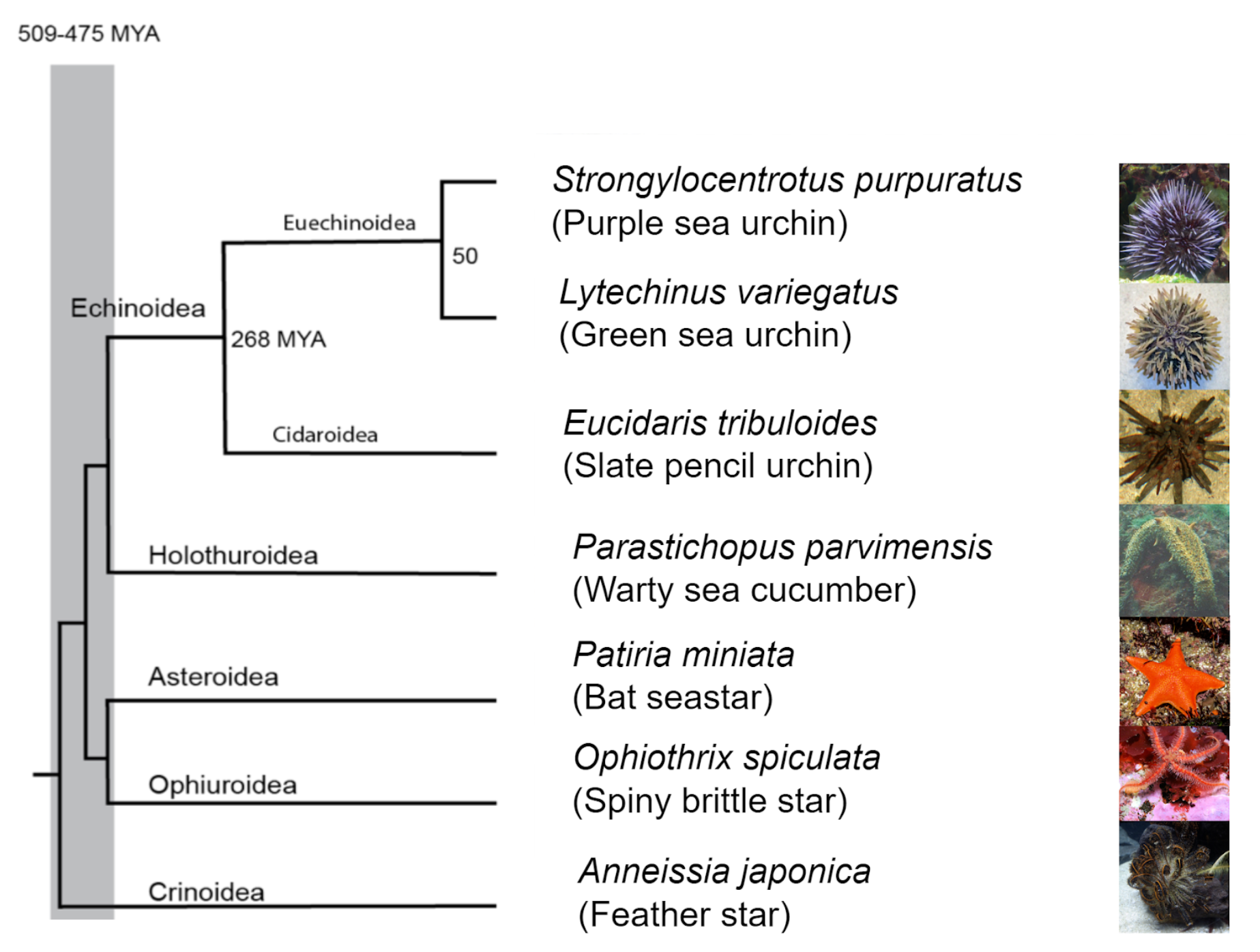
The placenta is a temporary organ that facilitates the exchange of nutrients and gases between a mother and a developing fetus. Having emerged around 160 million years ago, the placenta has since diversified across many mammals, and has even arisen independently in other vertebrates, including some snakes and live-bearing fish. Along with its evolutionary diversity, the placenta presents astounding morphological diversity too, showing diverse auxiliary functions in different species. The marked functional diversity of the placenta, along with its recent evolutionary origin, make it a unique model for studying the genetic basis for evolutionary novelty. Over the course of my 9 weeks at the Crick, I used bioinformatic tools to evaluate gene family expansions across mammalian clades. I then triangulated this analysis with in-house expression data from the placenta and decidua, the part of the endometrium that undergoes pregnancy-specific modifications.
To begin, I took annotated genomes from Ensembl, breaking each protein-coding gene into Pfam domains, or domains of function. I arranged these genomes based on sequence order within the gene to create domain architectures that more accurately capture the function of genes. The end goal of this process was to cluster genes into functional families. Expansions in each of these families were detected by comparing gene copy numbers corresponding to these families between mammalian clades using pairwise statistical tests.
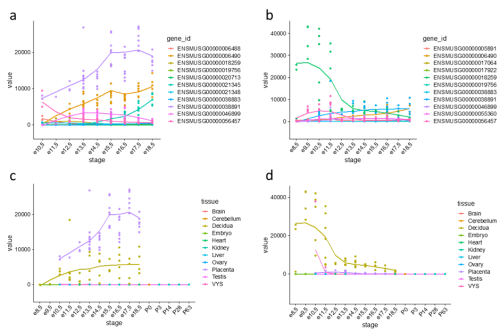
Figure 1: Expression trajectories of PRL (prolactins) in mouse. (a) Trajectories of 11 PRL genes in the placenta. (b) Trajectories of 11 PRL genes in the decidua. (c) Expression trajectory of Prl3b1 across all sampled tissues. (d) Expression trajectory of Prl8a2 across all sampled tissues.
One gene family expansion observed was that of rodent-specific prolactins. Prolactins in humans are released by the pituitary gland and are multifunctional throughout pregnancy, controlling key growth processes and lactation. On average, however, an additional 20 prolactin-domain-containing genes were observed in rodent species when compared to primates. Figures 1(a) and 1(b) highlight the expression trajectories of prolactins in the placenta and decidua in mice; clearly, prolactins are very highly expressed in both tissues (significant expression shown by values >1). However, the expression profiles differ slightly in that highly expressed prolactins in the placenta increase in expression through developmental time, while the inverse occurs in the decidua. Figures 1(c) and 1(d) show the most highly expressed genes in the placenta and decidua, respectively, shown in all tissues instead to highlight that this extreme expression is limited to the the placenta and decidua. Ben-Jonathan et al. (2008) reviews a plethora of evidence that suggests an important role of prolactins in rodents; for example, prolactin expression is key for downregulating interleukin expression and upregulating estrogen receptor expression, both required for fetus survival in rats. The role of the expansion of prolactins and their connection to a specific placental phenotype, however, remains unexplored.
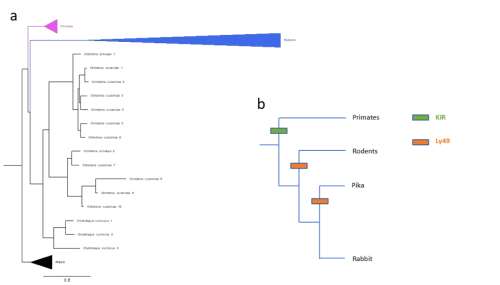
Figure 2: (a) Gene tree of Ly49 receptors across mammals, including main clades along with pig, horse, bison, sheep, and cow (abbreviated as PHACS). (b) Schematic of KIR/Ly49 expansion according to species tree.
Importantly, these pairwise statistical tests were unbiased in the sense that they included all significant gene family expansions, not only those pre-screened to be relevant to pregnancy. As such, for further analyses, I narrowed my field of search to revolve around genes already known to be relevant to placenta development and pregnancy, in this case natural killer cell receptors. Slodkowicz and Goldman (2020) showed that positive selection occurs more frequently in genes that carry immune functions. Additionally, uterine natural killer cells (uNKs) perform important functions throughout pregnancy, such as vascular remodelling, through MHC-interacting receptors. Though mammals share some uNK cell receptors, there are two main families of variable receptors: Ly49 receptors are expanded in rodents, while killer immunoglobulin-like receptors (KIR) are expanded in primates and other mammals (McQueen et al., 2002). The status of these receptors in lagomorphs, a group of mammals closely related to rodents that includes rabbits and hares, is not well studied. To detect expansions in lagomorphs, I used OrthoFinder (Emms and Kelly, 2019) to cluster genes by sequence, accounting for phylogenetic differences, screening for clusters of NK cell receptors. Figure 2(a) shows a phylogeny constructed of a cluster of Ly49 receptors; as expected, primates possess single copies of the receptor while rodents show a large expansion. However, two lagomorph species of pika show a late expansion of Ly49 receptors, while rabbits do not show the same. No compensatory expansion of KIRs was detected in rabbits either, an observation cross-referenced by searching the genomes of 3 closely related species (snowshoe hare, mountain hare, and brush rabbit). Instead, an expansion of leukocyte immunoglobulin-like receptors (LILRs) was detected. My project ended with testing de-novo genome annotation as a way of further elucidating what receptors rabbits and closely related species may be using.
Taken together, my time at the Crick generated data that provides various possible avenues for exploration, including investigating expansions that may underly interesting placental phenotypes, or better understanding immune genes during pregnancy in understudied species. Personally, I took home important skills in analyzing data, comparing methods, and substantially improved my working knowledge of programming languages like R and Python. I would like to thank my supervisor, Greg Slodkowicz, and Margarida Cardoso-Moreira for their friendly guidance throughout the project. I am extremely grateful to the Crick and the Rosa Beddington Fund for providing me with this unforgettable opportunity. I encourage all eligible students to apply for any future opportunities, as this has been an incredibly unique and gratifying experience.
If you are interested, please see the lab’s page for further details. You can contact me via Twitter.


 (10 votes)
(10 votes)

 (No Ratings Yet)
(No Ratings Yet)