June in preprints
Posted by the Node, on 6 July 2021
Welcome to our monthly trawl for developmental biology (and related) preprints.
The preprints this month are hosted on bioRxiv, arXiv and preprints.org – use these links to get to the section you want.
- Patterning & signalling
- Morphogenesis & mechanics
- Genes & genomes
- Stem cells, regeneration & disease modelling
- Plant development
- Evo-devo
Developmental biology
| Patterning & signalling
Ssdp influences Wg expression and embryonic somatic muscle identity in Drosophila melanogaster
Preethi Poovathumkadavil, Jean-Philippe Da Ponte, Krzysztof Jagla
Inhibition of aryl hydrocarbon receptor signaling promotes the terminal differentiation of human erythrocytes
Yijin Chen, Yong Dong, Xulin Lu, Wanjing Li, Yimeng Zhang, Bin Mao, Xu Pan, Xiaohong Li, Ya Zhou, Quanming An, Fangxin Xie, Shihui Wang, Yuan Xue, Xinping Cai, Mowen Lai, Qiongxiu Zhou, Yan Yan, Ruohan Fu, Hong Wang, Tatsutoshi Nakahata, Xiuli An, Lihong Shi, Yonggang Zhang, Feng Ma
Insulin is expressed by enteroendocrine cells during human fetal development
Adi Egozi, Dhana Llivichuzhca-Loja, Blake McCourt, Lydia Farack, Xiaojing An, Fujing Wang, Kong Chen, Liza Konnikova, Shalev Itzkovitz
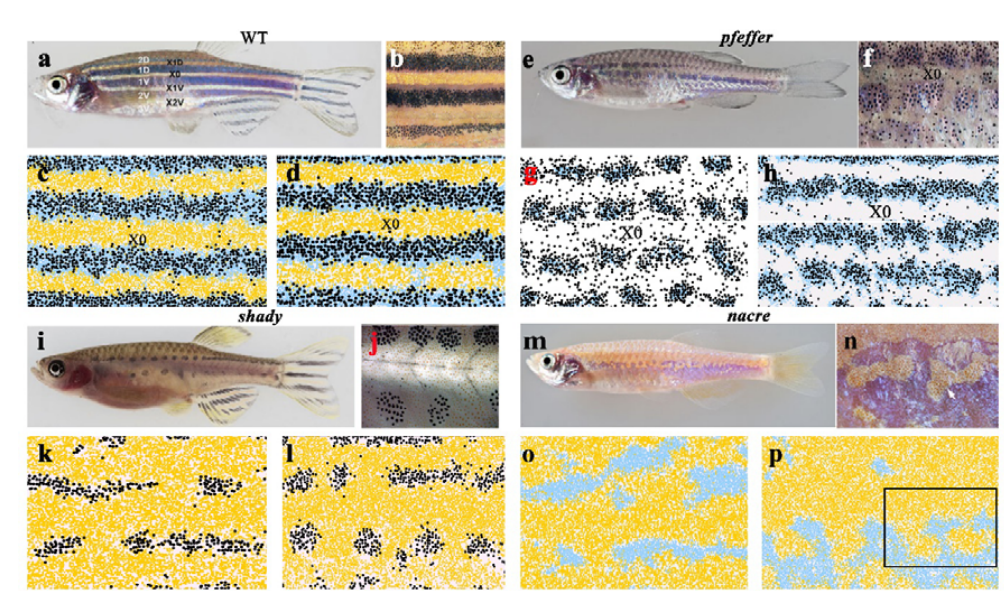
Differential growth is a critical determinant of zebrafish pigment pattern formation
Jennifer P. Owen, Christian A. Yates, Robert N. Kelsh
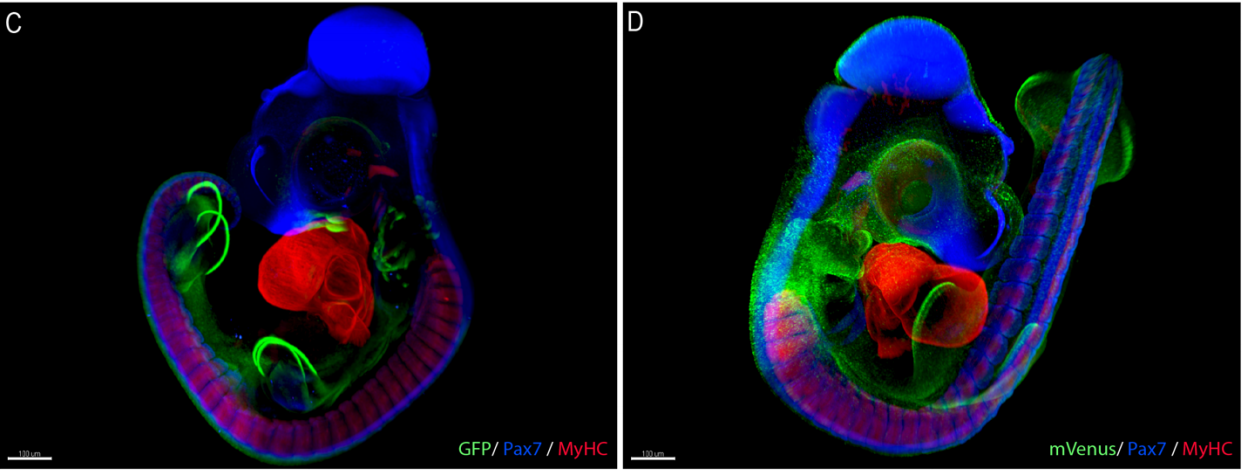
TopFlash transgenic quail reveals dynamic TCF/β-catenin signaling during avian embryonic development
Hila Barzilai-Tutsch, Valerie Morin, Gauthier Toulouse, Stephen Firth, Christophe Marcelle, Olivier Serralbo
In vitro endoderm emergence and self-organisation in the absence of extraembryonic tissues and embryonic architecture
Stefano Vianello, Matthias P. Lutolf
Serotonergic neuron ribosomes regulate the neuroendocrine control of Drosophila development
Lisa P. Deliu, Deeshpaul Jadir, Abhishek Ghosh, Savraj S. Grewal
Differential repression of Otx2 underlies the capacity of NANOG and ESRRB to induce germline entry
Matúš Vojtek, Jingchao Zhang, Juanjuan Sun, Man Zhang, Ian Chambers
The ciliary gene INPP5E confers dorsal telencephalic identity to human cortical organoids by negatively regulating Sonic Hedgehog signalling
Leah Schembs, Ariane Willems, Kerstin Hasenpusch-Theil, James D. Cooper, Katie Whiting, Karen Burr, Sunniva M.K. Bøstrand, Bhuvaneish T. Selvaraj, Siddharthan Chandran, Thomas Theil
Naa12 compensates for Naa10 in mice in the amino-terminal acetylation pathway
Hyae Yon Kweon, Mi-Ni Lee, Max Dörfel, Seungwoon Seo, Leah Gottlieb, Thomas Papazyan, Nina McTiernan, Rasmus Ree, David Bolton, Andrew Garcia, Michael Flory, Jonathan Crain, Alison Sebold, Scott Lyons, Ahmed Ismail, Elaine Marchi, Seong-keun Sonn, Se-Jin Jeong, Sejin Jeon, Shinyeong Ju, Simon J. Conway, TaeSoo Kim, Hyun-Seok Kim, Cheolju Lee, Tae-Young Roh, Thomas Arnesen, Ronen Marmorstein, Goo Taeg Oh, Gholson J. Lyon
Developmental Regulation of Homeostatic Plasticity in Mouse Primary Visual Cortex
Wei Wen, Gina G. Turrigiano
Patterning on the move: the effects of Hh morphogen source movement on signaling dynamics
D. G. Míguez, A. Iannini, D. García-Morales, F. Casares
Preterm birth alters the development of cortical microstructure and morphology at term-equivalent age
Ralica Dimitrova, Maximilian Pietsch, Judit Ciarrusta, Sean P. Fitzgibbon, Logan Z. J. Williams, Daan Christiaens, Lucilio Cordero-Grande, Dafnis Batalle, Antonios Makropoulos, Andreas Schuh, Anthony N. Price, Jana Hutter, Rui PAG Teixeira, Emer Hughes, Andrew Chew, Shona Falconer, Olivia Carney, Alexia Egloff, J-Donald Tournier, Grainne McAlonan, Mary A. Rutherford, Serena J. Counsell, Emma C. Robinson, Joseph V. Hajnal, Daniel Rueckert, A. David Edwards, Jonathan O’Muircheartaigh
Connectomes across development reveal principles of brain maturation
Daniel Witvliet, Ben Mulcahy, James K. Mitchell, Yaron Meirovitch, Daniel R. Berger, Yuelong Wu, Yufang Liu, Wan Xian Koh, Rajeev Parvathala, Douglas Holmyard, Richard L. Schalek, Nir Shavit, Andrew D. Chisholm, Jeff W. Lichtman, Aravinthan D.T. Samuel, Mei Zhen
Homothorax Controls a Binary Rhodopsin Switch in Drosophila Ocelli
Abhishek Kumar Mishra, Cornelia Fritsch, Roumen Voutev, Richard S. Mann, Simon G. Sprecher
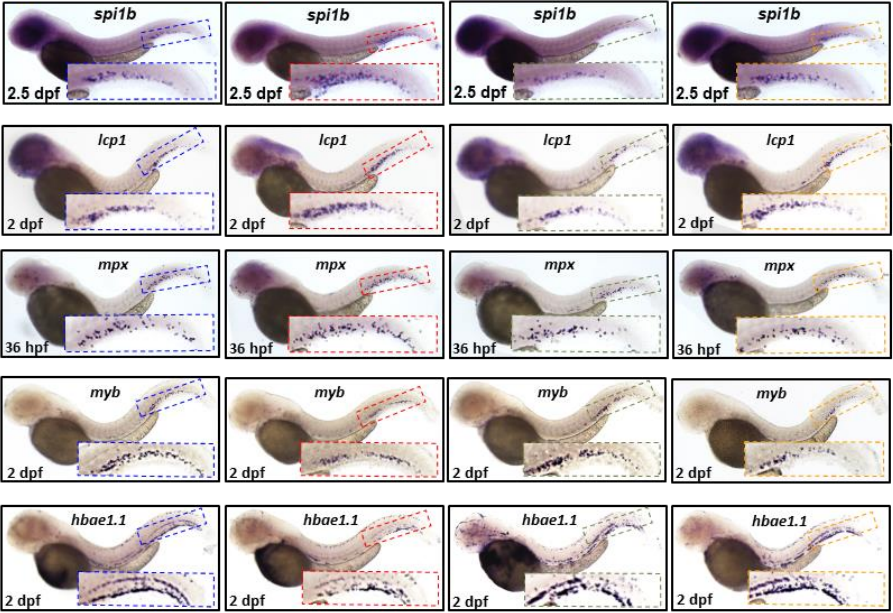
Genetic and chemical inhibition of autophagy in zebrafish induced myeloproliferation
Kazi Md Mahmudul Hasan, Xiang-Ke Chen, Zhen-Ni Yi, Jack Jark-Yin Lau, Alvin Chun-hang Ma
The asymmetric Pitx2 regulates intestinal muscular-lacteal development and protects against fatty liver disease
Shing Hu, Aparna Mahadevan, Isaac F. Elysee, Joseph Choi, Nathan R. Souchet, Gloria H. Bae, Alessandra K. Taboada, Gerald E. Duhamel, Carolyn S. Sevier, Ge Tao, Natasza A. Kurpios
The atypical RNA-binding protein TAF15 regulates dorsoanterior neural development through diverse mechanisms in Xenopus tropicalis
Caitlin S. DeJong, Darwin S. Dichmann, Cameron R. T. Exner, Yuxiao Xu, Richard M. Harland
Estrogen regulates early embryonic development of the olfactory sensory system via estrogen-responsive glia
Aya Takesono, Paula Schirrmacher, Aaron Scott, Jon M. Green, Okhyun Lee, Matthew J. Winter, Tetsuhiro Kudoh, Charles R. Tyler
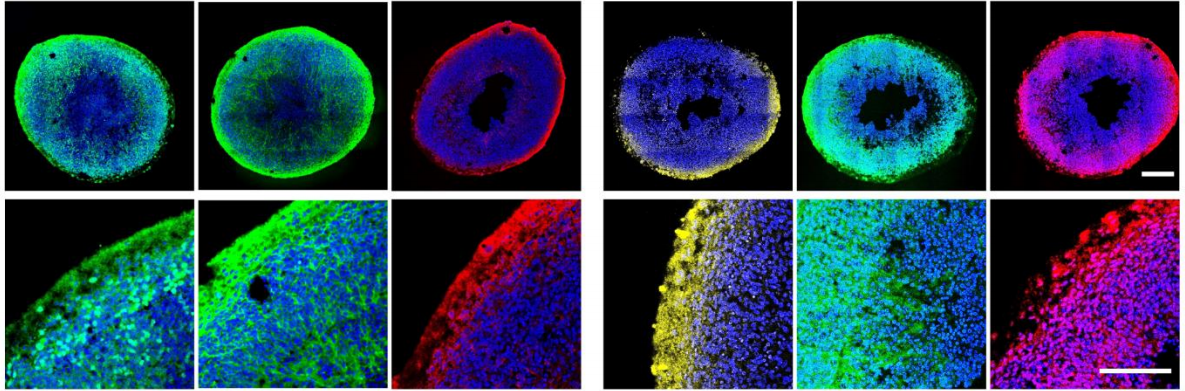
Chronic opioid treatment arrests neurodevelopment and alters synaptic activity in human midbrain organoids
Hye Sung Kim, Yang Xiao, Xuejing Chen, Siyu He, Jongwon Im, Moshe J. Willner, Michael O. Finlayson, Cong Xu, Huixiang Zhu, Se Joon Choi, Eugene V. Mosharov, Hae-Won Kim, Bin Xu, Kam W. Leong
Polypeptides IGF-1C and P24 synergistically promote osteogenic differentiation of bone marrow mesenchymal stem cells in vitro through the p38 and JNK signaling pathways
Gaoying Ran, Wei Fang, Lifang Zhang, Yuting Peng, Jiatong Li, Xianglong Ding, Shuguang Zeng, Yan He
Amyloid precursor protein localises to ependymal cilia in vertebrates and is required for ciliogenesis and brain development in zebrafish
Jasmine Chebli, Maryam Rahmati, Tammaryn Lashley, Birgitta Edeman, Anders Oldfors, Henrik Zetterberg, Alexandra Abramsson
Notch pathway is required for protection against heat-stress in spermatogonial stem cells
Omar D. Moreno Acosta, Agustín F. Boan, Ricardo S. Hattori, Juan I. Fernandino
In vivo imaging of mammary epithelial cell dynamics in response to lineage-biased Wnt/β-catenin activation
Bethan Lloyd-Lewis, Francesca Gobbo, Meghan Perkins, Guillaume Jacquemin, Marisa M Faraldo, Silvia Fre
Apoptotic Find-me Signals are an Essential Driver of Stem Cell Conversion To The Cardiac Lineage
Loic Fort, Vivian Gama, Ian G. Macara
Notch-dependent Abl signaling regulates cell motility during ommatidial rotation in Drosophila
Yildiz Koca, Linh T. Vuong, Jaskirat Singh, Edward Giniger, Marek Mlodzik
EXC-4/CLIC, Gα, and Rho/Rac signaling regulate tubulogenesis in C. elegans
Anthony F. Arena, Daniel D. Shaye
Release of Notch activity coordinated by IL-1β signalling confers differentiation plasticity of airway progenitors via Fosl2 during alveolar regeneration
Jinwook Choi, Yu Jin Jang, Catherine Dabrowska, Elhadi Iich, Kelly V. Evans, Helen Hall, Sam M. Janes, Benjamin D. Simons, Bon-Kyoung Koo, Jonghwan Kim, Joo-Hyeon Lee
Reciprocal EGFR signaling in the Anchor Cell ensures precise inter-organ connection during C. elegans vulval morphogenesis
Silvan Spiri, Simon Berger, Louisa Mereu, Andrew DeMello, Alex Hajnal
Synergistic TOR and ERK inhibition mitigates the hereditary haemorrhagic telangiectasia-like phenotype and excess kugel formation in endoglin mutant zebrafish
Ryan O. Snodgrass, Helen M. Arthur, Timothy J.A. Chico
Gene-teratogen interactions influence the penetrance of birth defects by altering Hedgehog signaling strength
Jennifer H. Kong, Cullen B. Young, Ganesh V. Pusapati, F Hernán Espinoza, Chandni B. Patel, Francis Beckert, Sebastian Ho, Bhaven B. Patel, George C. Gabriel, L. Aravind, J Fernando Bazan, Teresa M. Gunn, Cecilia W. Lo, Rajat Rohatgi
GLI transcriptional repression is inert prior to Hedgehog pathway activation
Rachel K. Lex, Weiqiang Zhou, Zhicheng Ji, Kristin N. Falkenstein, Kaleigh E. Schuler, Kathryn E. Windsor, Joseph D. Kim, Hongkai Ji, Steven A Vokes
FGF/ERK autocrine signaling is enhanced by NANOG in a subpopulation of pluripotent stem cells to execute autoregulation and induce heterogeneity
Hanuman T Kale, Rajendra Singh Rajpurohit, Debabrata Jana, Vishnu V Vijay, Mansi Srivastava, Preeti R Mourya, Gunda Srinivas, P Chandra Shekar
Neuronal KGB-1 JNK MAPK signaling regulates the dauer developmental decision in response to environmental stress in C. elegans
Deepshikha Dogra, Warakorn Kulalert, Frank C. Schroeder, Dennis H. Kim
Self-Organogenesis from 2D Micropatterns to 3D Biomimetic Biliary Trees
Emilie Gontran, Lorena Loarca, Cyrille El Khassis, Latifa Bouzhir, Dmitry Ayollo, Elsa Mazari-Arrighi, Alexandra Fuchs, Pascale Dupuis-Williams
| Morphogenesis & mechanics
Differential adhesion regulates neurite placement via a retrograde zippering mechanism
Titas Sengupta, Noelle L. Koonce, Mark W. Moyle, Leighton H. Duncan, Nabor Vázquez-Martínez, Sarah E. Emerson, Xiaofei Han, Lin Shao, Yicong Wu, Anthony Santella, Li Fan, Zhirong Bao, William A. Mohler, Hari Shroff, Daniel A. Colón-Ramos
Acetylated microtubules are required for maintenance of the barrier between two adjacent tissues
Matthew Antel, Taylor Simao, Muhammed Burak Bener, Mayu Inaba
A midbody component homolog, too much information/prc1-like, is required for microtubule reorganization during both cytokinesis and axis induction in the early zebrafish embryo
S Nair, E.L. Welch, C.E. Moravec, R.L. Trevena, F. Pelegri
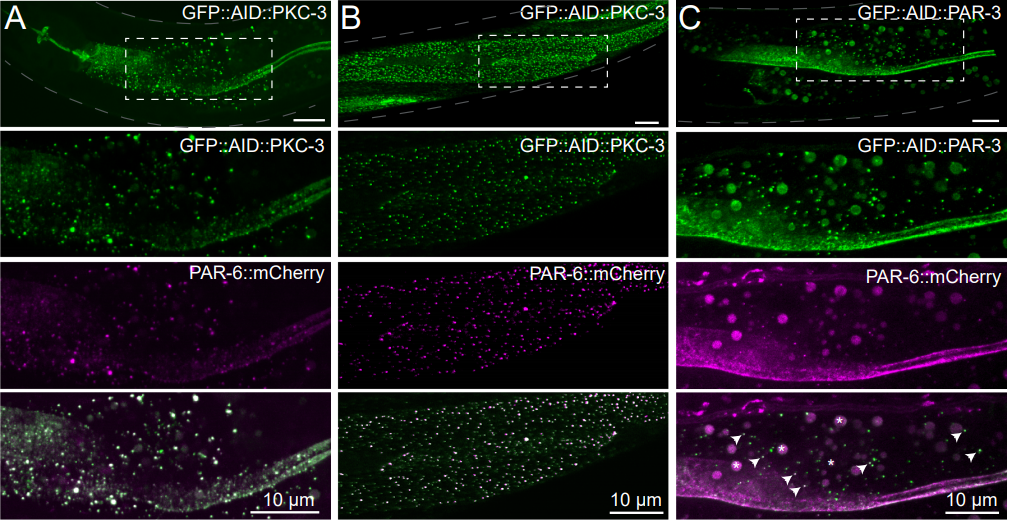
Pak1 and PP2A antagonize aPKC function to support cortical tension induced by the Crumbs-Yurt complex
Cornélia Biehler, Katheryn E. Rothenberg, Alexandra Jetté, Hélori-Mael Gaudé, Rodrigo Fernandez-Gonzalez, Patrick Laprise
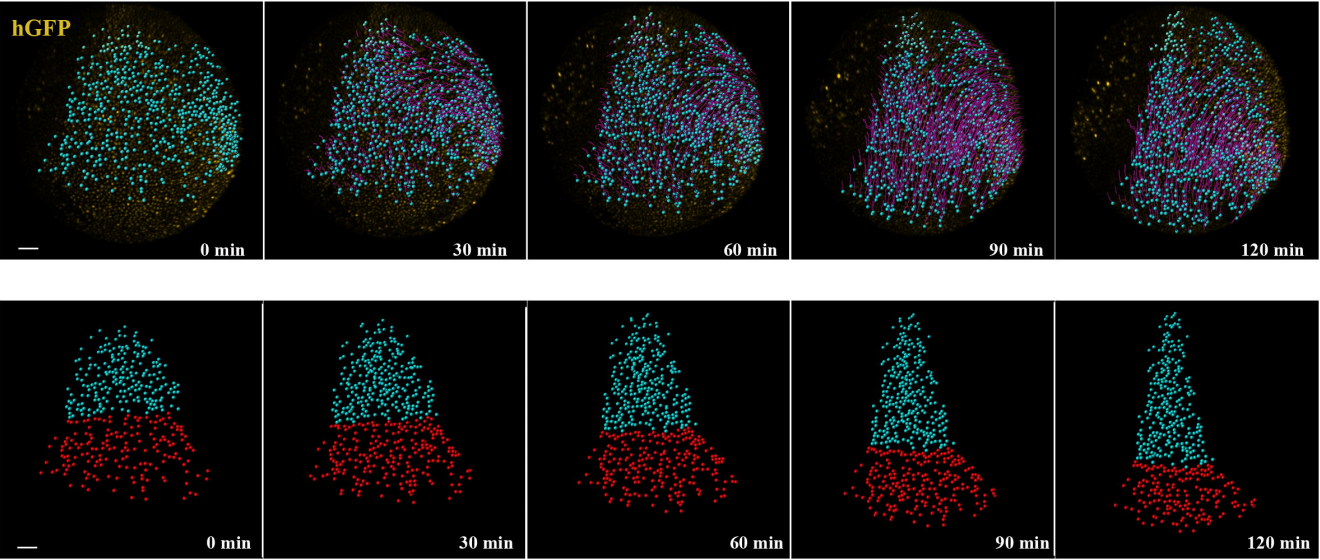
Distinct spatiotemporal contribution of morphogenetic events and mechanical tissue coupling during Xenopus neural tube closure
Neophytos Christodoulou, Paris A. Skourides
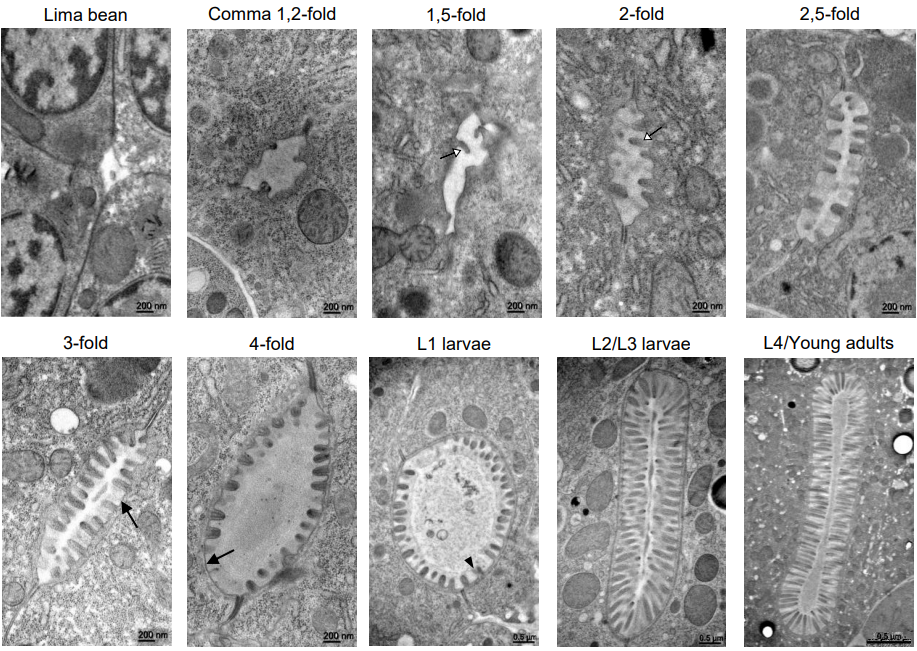
High resolution dynamic mapping of the C. elegans intestinal brush border
Aurélien Bidaud-Meynard, Flora Demouchy, Ophélie Nicolle, Anne Pacquelet, Grégoire Michaux
Met-signaling Controls Dendritic Cell Migration by Regulating Podosome Formation and Function
Ahmed E.I. Hamouda, Carmen Schalla, Antonio Sechi, Martin Zenke, Thomas Hieronymus
Cell-matrix adhesion contributes to permeability control in human colon organoids
James Varani, Shannon D. McClintock, Muhammad N. Aslam
Cell adhesions link subcellular actomyosin dynamics to tissue scale force production during vertebrate convergent extension
Robert J. Huebner, Shinuo Weng, Chanjae Lee, Sena Sarıkaya, Ophelia Papoulas, Rachael M. Cox, Edward M. Marcotte, John B. Wallingford
Convergent extension requires adhesion-dependent biomechanical integration of cell crawling and junction contraction
Shinuo Weng, Robert J. Huebner, John B. Wallingford
A new approach to measure forces at junction vertices in an epithelium
Clémentine Villeneuve, Samuel Mathieu, Emilie Lagoutte, Bruno Goud, Philippe Chavrier, Jean-Baptiste Manneville, Carine Rossé
Pressure and curvature control of contact inhibition in epithelia growing under spherical confinement
Ilaria Di Meglio, Anastasiya Trushko, Pau Guillamat, Carles Blanch-Mercader, Aurélien Roux
Mechanical Stimulation via Muscle Activity is Necessary for the Maturation of Tendon Multiscale Mechanics during Embryonic Development
Benjamin E Peterson, Rebecca A. Rolfe, Allen Kunselman, Paula Murphy, Spencer E. Szczesny
Laminin-binding Integrins Regulate Angiogenesis by Distinct and Overlapping Mechanisms in Organotypic Cell Culture Models
Hao Xu, Susan E LaFlamme
Bmper is required for morphogenesis of the anterior and posterior semicircular canal ducts in the developing zebrafish inner ear
Sarah Baxendale, Esther C. Maier, Nikolaus D. Obholzer, Sarah Burbridge, Joseph Zinski, Francesca B. Tuazon, Nicholas J. van Hateren, M. Montserrat Garcia Romero, Mar Marzo, Kazutomo Yokoya, Robert D. Knight, Sean G. Megason, Mary C. Mullins, Tanya T. Whitfield
Characterisation of the transcriptional dynamics underpinning the function, fate, and migration of the mouse Anterior Visceral Endoderm
Shifaan Thowfeequ, Jonathan Fiorentino, Di Hu, Maria Solovey, Sharon Ruane, Maria Whitehead, Bart Vanhaesebroeck, Antonio Scialdone, Shankar Srinivas
IFT20 is critical for early chondrogenesis during endochondral ossification
Hiroyuki Yamaguchi, Megumi Kitami, Karin H. Uchima Koecklin, Li He, Jianbo Wang, Daniel S. Perrien, William R. Lagor, Yoshihiro Komatsu
| Genes & genomes
Caenorhabditis elegans ETR-1/CELF has broad effects on the muscle cell transcriptome, including genes that regulate translation and neuroblast migration
Matthew E. Ochs, Rebecca McWhirter, Rob Unckless, David Miller, Erik A Lundquist
A comprehensive series of temporal transcription factors in the fly visual system
Nikolaos Konstantinides, Anthony M. Rossi, Aristides Escobar, Liébaut Dudragne, Yen-Chung Chen, Thinh Tran, Azalia Martinez Jaimes, Mehmet Neset Özel, Félix Simon, Zhiping Shao, Nadejda M. Tsankova, John F. Fullard, Uwe Walldorf, Panos Roussos, Claude Desplan
Expansion of RNA sequence diversity and RNA editing rates throughout human cortical development
Ryn Cuddleston, Laura Sloofman, Lindsay Liang, Enrico Mossotto, Xuanjia Fan, Minghui Wang, Bin Zhang, Jiebiao Wang, Nenad Sestan, Bernie Devlin, Kathryn Roeder, Joseph D. Buxbaum, Stephan J. Sanders, Michael S. Breen
AP-2α and AP-2β cooperatively function in the craniofacial surface ectoderm to regulate chromatin and gene expression dynamics during facial development
Eric Van Otterloo, Isaac Milanda, Hamish Pike, Hong Li, Kenneth L Jones, Trevor Williams
A comprehensive temporal patterning gene network in Drosophila medulla neuroblasts revealed by single-cell RNA sequencing
Hailun Zhu, Sihai Dave Zhao, Alokananda Ray, Yu Zhang, Xin Li
Heterogeneity and molecular programming of progenitors for motor neurons and oligodendrocytes
Lingyan Xing, Rui Chai, Jiaqi Wang, Jiaqi Lin, Hanyang Li, Yueqi Wang, Biqin Lai, Junjie Sun, Gang Chen
A DNA Replication-Independent Function of the pre-Replication Complex during Cell Invasion in C. elegans
Evelyn Lattmann, Ting Deng, Michael Walser, Patrizia Widmer, Charlotte Rexha-Lambert, Vibhu Prasad, Ossia Eichhoff, Michael Daube, Reinhard Dummer, Mitchell P. Levesque, Alex Hajnal
CDK12 is Necessary to Promote Epidermal Differentiation through Transcription Elongation
Jingting Li, Manisha Tiwari, Yifang Chen, George L. Sen
Systematic reconstruction of the cellular trajectories of mammalian embryogenesis
Chengxiang Qiu, Junyue Cao, Tony Li, Sanjay Srivatsan, Xingfan Huang, Diego Calderon, William Stafford Noble, Christine M. Disteche, Malte Spielmann, Cecilia B. Moens, Cole Trapnell, Jay Shendure
Identification of enamel knot gene signature within the developing mouse molar
Emma Wentworth Winchester, Justin Cotney
Spatial cell type mapping of the oligodendrocyte lineage in the mouse juvenile and adult CNS with in situ sequencing
Markus M. Hilscher, Christoffer Mattsson Langseth, Petra Kukanja, Chika Yokota, Mats Nilsson, Gonçalo Castelo-Branco
Application of ATAC-Seq for genome-wide analysis of the chromatin state at single myofiber resolution
Korin Sahinyan, Darren M Blackburn, Marie-Michelle Simon, Felicia Lazure, Tony Kwan, Guillaume Bourque, Vahab D Soleimani
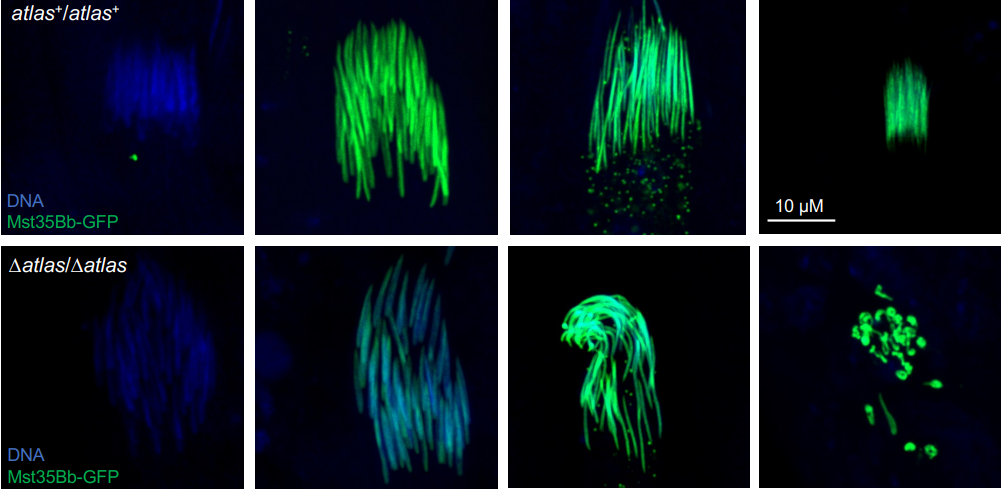
A putative de novo evolved gene required for spermatid chromatin condensation in Drosophila melanogaster
Emily L. Rivard, Andrew G. Ludwig, Prajal H. Patel, Anna Grandchamp, Sarah E. Arnold, Alina Berger, Emilie M. Scott, Brendan J. Kelly, Grace C. Mascha, Erich Bornberg-Bauer, Geoffrey D. Findlay
Klf5 establishes bi-potential cell fate by dual regulation of ICM and TE specification genes
Martin Kinisu, Yong Jin Choi, Claudia Cattoglio, Ke Liu, Hector Roux de Bezieux, Raeline Valbuena, Nicole Pum, Sandrine Dudoit, Haiyan Huang, Zhenyu Xuan, Sang Yong Kim, Lin He
C. elegans TFIIH subunit GTF-2H5/TTDA is a non-essential transcription factor indispensable for DNA repair
Karen L. Thijssen, Melanie van der Woude, Carlota Davó-Martínez, Mariangela Sabatella, Wim Vermeulen, Hannes Lans
Adult fibroblasts retain organ-specific transcriptomic identity
Elvira Forte, Mirana Ramialison, Hieu T. Nim, Madison Mara, Rachel Cohn, Sandra L. Daigle, Sarah Boyd, J. Travis Hinson, Mauro W. Costa, Nadia A. Rosenthal, Milena B. Furtado
Differential Bcd activation of two hunchback promoters emerges from unified kinetics of enhancer-promoter interaction
Jingyao Wang, Shihe Zhang, Hongfang Lu, Heng Xu
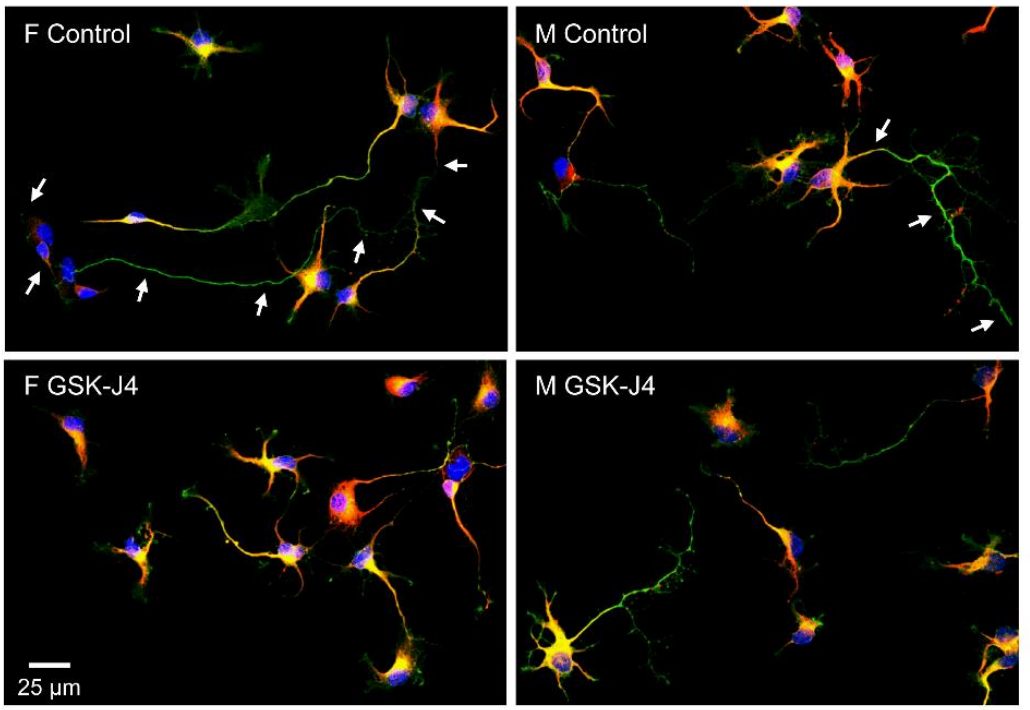
X-Linked Histone H3K27 Demethylase Kdm6a Regulates Sexually Dimorphic Differentiation of Hypothalamic Neurons
Lucas E. Cabrera Zapata, Carla D. Cisternas, Camila Sosa, Maria Angeles Arevalo, Luis Miguel Garcia-Segura, María Julia Cambiasso
Environment-driven reprogramming of gamete DNA methylation occurs during maturation and is transmitted intergenerationally in salmon
Kyle Wellband, David Roth, Tommi Linnansaari, R. Allen Curry, Louis Bernatchez
ETV2 primes hematoendothelial gene enhancers prior to hematoendothelial fate commitment
Jeffrey D. Steimle, Chul Kim, Rangarajan D. Nadadur, Zhezhen Wang, Andrew D. Hoffmann, Erika Hanson, Junghun Kweon, Tanvi Sinha, Kyunghee Choi, Brian L. Black, John M. Cunningham, Kohta Ikegami, Ivan P. Moskowitz
Multivariate genome-wide association studies on the tissue compartments of human brain identify novel loci underpinning brain development and neuropsychiatric outcomes
Chun Chieh Fan, Robert Loughnan, Carolina Makowski, Diliana Pechva, Chi-Hua Chen, Donald Hagler, Wesley K. Thompson, Dennis van der Meer, Oleksandr Frei, Ole Andreassen, Anders M. Dale
Chromatin dynamics during hematopoiesis reveal discrete regulatory modules instructing differentiation
Grigorios Georgolopoulos, Nikoletta Psatha, Mineo Iwata, Andrew Nishida, Tannishtha Som, Minas Yiangou, John A. Stamatoyannopoulos, Jeff Vierstra
H3K9 tri-methylation at Nanog times differentiation commitment and enables the acquisition of primitive endoderm fate
A. Dubois, L. Vincenti, A. Chervova, S. Vandormael-Pournin, M. Cohen-Tannoudji, P. Navarro
Deconvolution of the epigenetic age discloses distinct inter-personal variability in epigenetic aging patterns
Tamar Shahal, Elad Segev, Thomas Konstantinovsky, Yonit Marcus, Gabi Shefer, Metsada Pasmanik-Chor, Assaf Buch, Yuval Ebenstein, Paul Zimmet, Naftali Stern
The Caenorhabditis elegans TDRD5/7-like protein, LOTR-1, interacts with the helicase ZNFX-1 to balance epigenetic signals in the germline
Elisabeth A. Marnik, Miguel V. Almeida, P. Giselle Cipriani, George Chung, Edoardo Caspani, Emil Karaulanov, Falk Butter, Catherine S. Sharp, John Zinno, Hin Hark Gan, Fabio Piano, René F Ketting, Kristin C. Gunsalus, Dustin L. Updike
Blm Helicase Facilitates Rapid Replication of Repetitive DNA Sequences in early Drosophila Development
Jolee M. Ruchert, Morgan M Brady, Susan McMahan, Karly J. Lacey, Leigh C. Latta, Jeff Sekelsky, Eric P. Stoffregen
Translesion DNA synthesis-driven mutagenesis in very early embryogenesis of fast cleaving embryos
Elena Lo Furno, Isabelle Busseau, Claudio Lorenzi, Cima Saghira, Matt C Danzi, Stephan Zuchner, Domenico Maiorano
Identification of PAX6 and NFAT4 as the transcriptional regulators of lncRNA Mrhl in neuronal progenitors
Debosree Pal, Sangeeta Dutta, Dhanur P Iyer, Utsa Bhaduri, M.R.S Rao
Conserved Transcription Factors Control Chromatin Accessibility and Gene Expression to Maintain Cell Fate Stability and Restrict Reprogramming of Differentiated Cells
Maria A. Missinato, Sean A. Murphy, Michaela Lynott, Anaïs Kervadec, Michael S. Yu, Yu-Ling Chang, Suraj Kannan, Mafalda Loreti, Christopher Lee, Prashila Amatya, Hiroshi Tanaka, Chun-Teng Huang, Pier Lorenzo Puri, Chulan Kwon, Peter D. Adams, Li Qian, Alessandra Sacco, Peter Andersen, Alexandre R. Colas
ZFP462 targets heterochromatin to transposon-derived enhancers restricting transcription factor binding and expression of lineage-specifying genes
Ramesh Yelagandula, Karin Stecher, Maria Novatchkova, Luca Michetti, Georg Michlits, Jingkui Wang, Pablo Hofbauer, Carina Pribitzer, Gintautas Vainorius, Luke Isbel, Sasha Mendjan, Dirk Schübeler, Ulrich Elling, Julius Brennecke, Oliver Bell
MicroRNA-202 prevents precocious spermatogonial differentiation and meiotic initiation during mouse spermatogenesis
Jian Chen, Chenxu Gao, Xiwen Lin, Yan Ning, Wei He, Chunwei Zheng, Daoqin Zhang, Lin Yan, Binjie Jiang, Yuting Zhao, Md Alim Hossen, Chunsheng Han
RNAseq analysis reveals dynamic metaboloepigenetic profiles of human, mouse and bovine pre-implantation embryos
Marcella Pecora Milazzotto, Michael James Noonan, Marcia de Almeida Monteiro Melo Ferraz
| Stem cells, regeneration & disease modelling
The NF-κB pathway regulates heterochromatin at intronic young LINE-1 elements and hematopoietic stem cell gene expression during irradiation stress
Yanis Pelinski, Donia Hidaoui, François Hermetet, Anne Stolz, M’boyba Khadija Diop, Amir M. Chioukh, Françoise Porteu, Emilie Elvira-Matelot
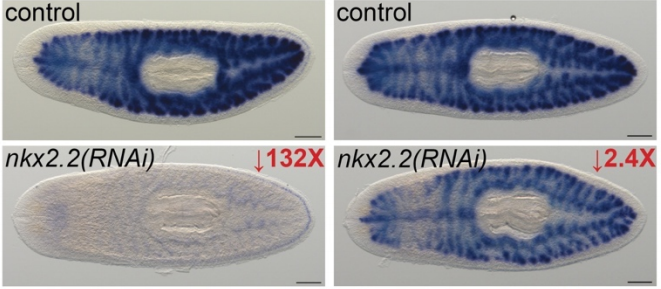
Intestine-enriched apolipoprotein b orthologs are required for stem cell differentiation and regeneration in planarians
Lily L. Wong, Christina G. Bruxvoort, Nicholas I. Cejda, Jannette Rodriguez Otero, David J. Forsthoefel
Proliferation maintains the undifferentiated status of stem cells: the role of the planarian cell cycle regulator Cdh1
Yuki Sato, Yoshihiko Umesono, Yoshihito Kuroki, Kiyokazu Agata, Chikara Hashimoto
Investigation of Thyroid Hormone Associated Gene-Regulatory Networks during Hepatogenesis using an Induced Pluripotent Stem Cell based Model
Audrey Ncube, Nina Graffmann, Jan Greulich, Bo Scherer, Wasco Wruck, James Adjaye
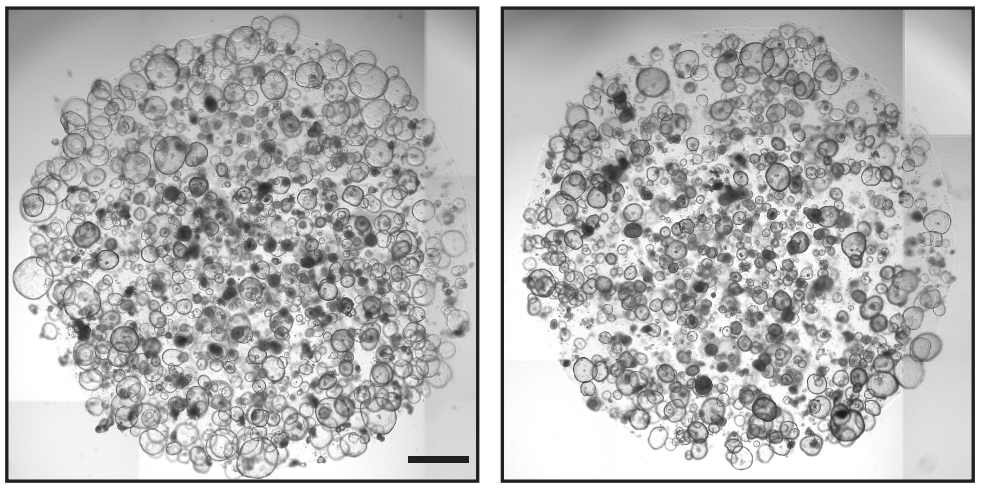
Robust differentiation of human enteroendocrine cells from intestinal stem cells
Daniel Zeve, Eric Stas, Joshua de Sousa Casal, Prabhath Mannam, Wanshu Qi, Xiaolei Yin, Sarah Dubois, Manasvi S. Shah, Erin P. Syverson, Sophie Hafner, Jeffrey M. Karp, Diana L. Carlone, Jose Ordovas-Montanes, David T. Breault
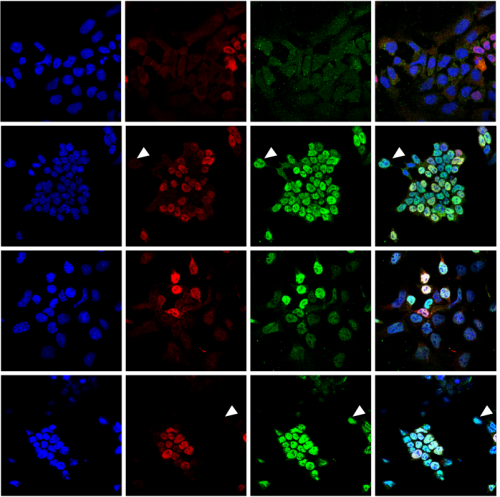
Loss of Resf1 reduces the efficiency of embryonic stem cell self-renewal and germline entry
Matúš Vojtek, Ian Chambers
Hypertrophic Chondrocytes Serve as a Reservoir for Unique Marrow Associated Skeletal Stem and Progenitor Cells, Osteoblasts, and Adipocytes During Skeletal Development
Jason T. Long, Abigail Leinroth, Yihan Liao, Yinshi Ren, Anthony J. Mirando, Tuyet Nguyen, Wendi Guo, Deepika Sharma, Colleen Wu, Kathryn Song Eng Cheah, Courtney M. Karner, Matthew J. Hilton
Identification of SUMO targets required to maintain human stem cells in the pluripotent state
Barbara Mojsa, Michael H. Tatham, Lindsay Davidson, Magda Liczmanska, Emma Branigan, Ronald T. Hay
Stem cell therapy for skin regeneration using mesenchymal stem cells derived from the progeroid Werner syndrome-specific iPS cells
Shinichiro Funayama, Hisaya Kato, Hiyori Kaneko, Kentaro Kosaka, Daisuke Sawada, Aki Takada-Watanabe, Takuya Minamizuka, Yusuke Baba, Masaya Koshizaka, Akira Shimamoto, Yasuo Ouchi, Atsushi Iwama, Yusuke Endo, Naoya Takayama, Koji Eto, Yoshiro Maezawa, Koutaro Yokote
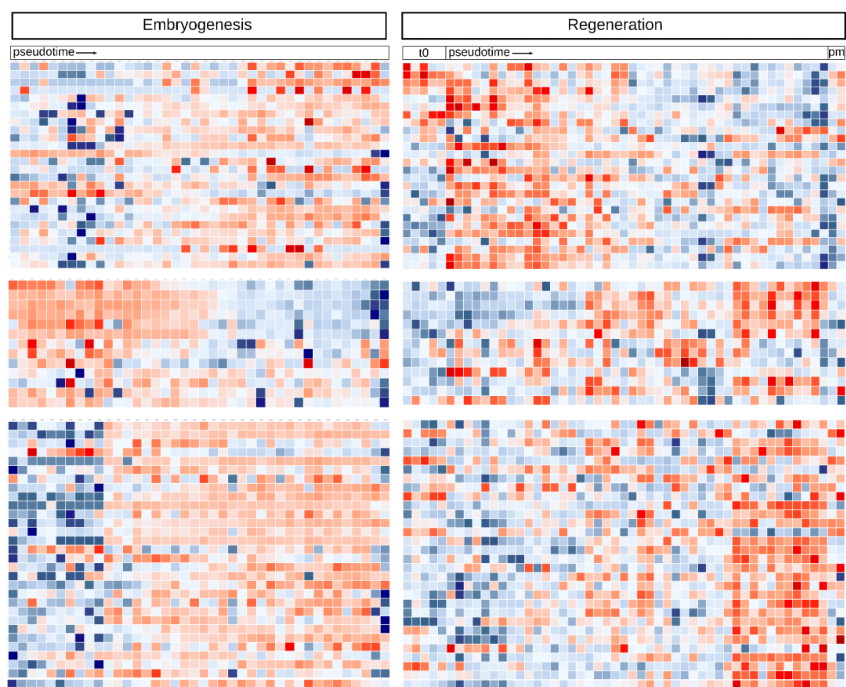
Distinct gene expression dynamics in developing and regenerating limbs
Chiara Sinigaglia, Alba Almazan, Marie Semon, Benjamin Gillet, Sandrine Hughes, Eric Edsinger, Michalis Averof, Mathilde Paris
Single cell chronoatlas of regenerating mouse livers reveals early Kupffer cell proliferation
Daniel Sánchez-Taltavull, Tess Brodie, Joel Zindel, Noëlle Dommann, Bas G.J. Surewaard, Adrian Keogh, Nicolas Mélin, Isabel Büchi, Riccardo Tombolini, Paul Kubes, Daniel Candinas, Guido Beldi, Deborah Stroka
Vestibular and auditory hair cell regeneration following targeted ablation of hair cells with diphtheria toxin in zebrafish
Erin Jimenez, Claire C. Slevin, Luis Colón-Cruz, Shawn M. Burgess
Notch signaling via Hey1 and Id2b regulates Müller glia’s regenerative response to retinal injury
Aresh Sahu, Sulochana Devi, Jonathan Jui, Daniel Goldman
Anal skin-like epithelium mediates colonic wound healing
Cambrian Y. Liu, Nandini Girish, Marie L. Gomez, Philip E. Dubé, M. Kay Washington, Benjamin D. Simons, D. Brent Polk
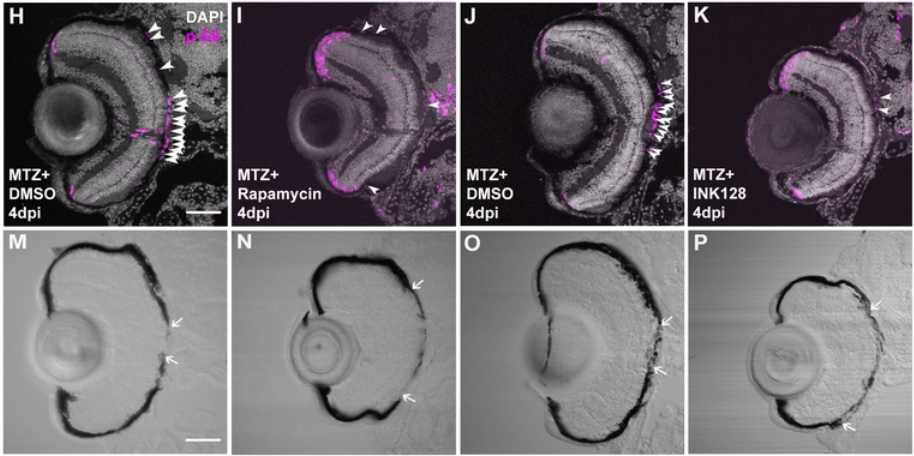
mTOR activity is essential for retinal pigment epithelium regeneration in zebrafish
Fangfang Lu, Lyndsay L. Leach, Jeffrey M. Gross
Potential therapy for progressive vision loss due to PCDH15-associated Usher Syndrome developed in an orthologous Usher mouse
Saumil Sethna, Wadih M. Zein, Sehar Riaz, Arnaud P. J. Giese, Julie M. Schultz, Todd Duncan, Robert B. Hufnagel, Carmen C. Brewer, Andrew J. Griffith, T. Michael Redmond, Saima Riazuddin, Thomas B. Friedman, Zubair M. Ahmed
A functional network signature in the developing cerebellum: evidence from a preclinical model of autism
María Berenice Soria-Ortiz, Atáulfo Martínez Torres, Daniel Reyes-Haro
Pharmacological inhibition of the VCP/proteasome axis rescues photoreceptor degeneration in RHOP23H rat retinal explants
Merve Sen, Oksana Kutsyr, Bowen Cao, Sylvia Bolz, Blanca Arango-Gonzalez, Marius Ueffing
Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome
Fikri Birey, Min-Yin Li, Aaron Gordon, Mayuri Thete, Alfredo M Valencia, Omer Revah, Anca M Pasca, Daniel H Geschwind, Sergiu P Pasca
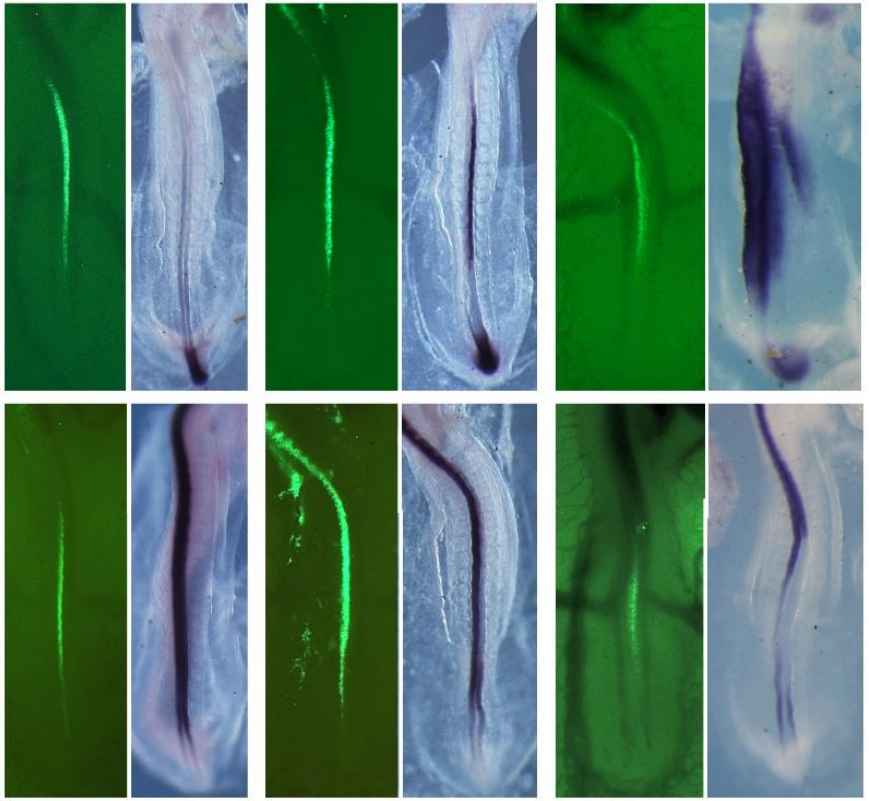
Sustained experimental activation of FGF8/ERK in the developing chicken spinal cord reproducibly models early events in ERK-mediated tumorigenesis
Axelle Wilmerding, Lauranne Bouteille, Nathalie Caruso, Ghislain Bidaut, Heather Etchevers, Yacine Graba, Marie-Claire Delfini
Mutations in SIX1 associated with Branchio-oto-renal Syndrome (BOR) differentially affect otic expression of putative target genes
Tanya Mehdizadeh, Himani Datta Majumdar, Sarah Ahsan, Andre Tavares, Sally A. Moody
Comparative therapeutic strategies for preventing aortic rupture in a mouse model of vascular Ehlers Danlos syndrome
Anne Legrand, Charline Guery, Julie Faugeroux, Erika Fontaine, Carole Beugnon, Amélie Gianfermi, Irmine Loisel-Ferreira, Marie-Christine Verpont, Salma Adham, Tristan Mirault, Juliette Hadchouel, Xavier Jeunemaitre
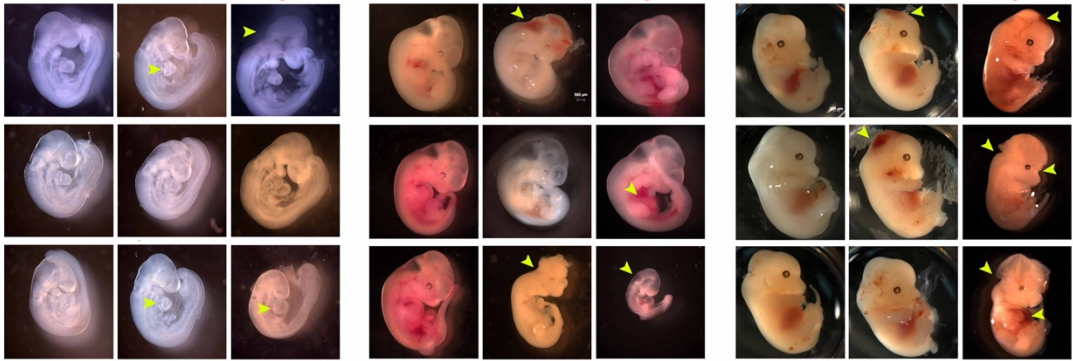
Maternal hyperglycemia impedes second heart field-derived cardiomyocyte differentiation to elevate the risk of congenital heart defects
Sathiyanarayanan Manivannan, Corrin Mansfield, Xinmin Zhang, Karthik M. Kodigepalli, Uddalak Majumdar, Vidu Garg, Madhumita Basu
Analysis of CHD-7 defective dauer nematodes implicates collagen misregulation in CHARGE syndrome features
Diego M. Jofré, Dane K. Hoffman, Ailen S. Cervino, McKenzie Grundy, Sijung Yun, Francis RG. Amrit, Donna B. Stolz, Esteban Salvatore, Fabiana A. Rossi, Arjumand Ghazi, M. Cecilia Cirio, Judith L. Yanowitz, Daniel Hochbaum
Identifying developing interneurons as a potential target for multiple genetic autism risk factors in human and rodent forebrain
Yifei Yang, Sam A. Booker, James M. Clegg, Idoia Quintana Urzainqui, Anna Sumera, Zrinko Kozic, Owen Dando, Sandra Martin Lorenzo, Yann Herault, Peter C. Kind, David J. Price, Thomas Pratt
BRN2 and PTN unveil multiple neurodevelopmental mechanisms in Schizophrenia patient-derived cerebral organoids
Michael Notaras, Aiman Lodhi, Friederike Dundar, Paul Collier, Nicole Sayles, Hagen Tilgner, David Greening, Dilek Colak
Inter- and intrapopulational heterogeneity of characteristic markers in adult human neural crest-derived stem cells
Beatrice A. Windmöller, Anna L. Höving, Johannes F.W. Greiner
PpRPK2 modulates auxin homeostasis and transport to specify stem cell identity and plant shape in the moss Physcomitrella
Zoe Nemec Venza, Connor Madden, Amy Stewart, Wei Liu, Ondřej Novák, Aleš Pěnčík, Andrew C. Cuming, Yasuko Kamisugi, C. Jill Harrison
Fetal-like reversion in the regenerating intestine is regulated by mesenchymal Asporin
Sharif Iqbal, Simon Andersson, Ernesta Nestaite, Nalle Pentinmikko, Ashish Kumar, Daniel Borshagovski, Anna Webb, Tuure Saarinen, Anne Juuti, Alessandro Ori, Markku Varjosalo, Kirsi H. Pietiläinen, Kim B. Jensen, Menno Oudhoff, Pekka Katajisto
A single-cell atlas of de novo β-cell regeneration reveals the contribution of hybrid β/δ cells to diabetes recovery in zebrafish
Sumeet Pal Singh, Prateek Chawla, Alisa Hnatiuk, Margrit Kamel, Luis Delgadillo Silva, Bastiaan Spanjard, Sema Elif Eski, Sharan Janjuha, Pedro Olivares, Oezge Kayisoglu, Fabian Rost, Juliane Bläsche, Annekathrin Kränkel, Andreas Petzold, Thomas Kurth, Susanne Reinhardt, Jan Philipp Junker, Nikolay Ninov
Modular, Cascade-like Transcriptional Program of Regeneration in Stentor
Pranidhi Sood, Athena Lin, Rebecca McGillivary, Wallace F. Marshall
A multimodal iPSC platform for cystic fibrosis drug testing
Andrew Berical, Rhianna E. Lee, Junjie Lu, Mary Lou Beermann, Jake A. LeSeur, Aditya Mithal, Dylan Thomas, Nicole Ranallo, Megan Peasley, Alex Stuffer, Jan Harrington, Kevin Coote, Killian Hurley, Paul McNally, Gustavo Mostovslavsky, John Mahoney, Scott H. Randell, Finn J. Hawkins
Dystonia-specific mutations in THAP1 alter transcription of genes associated with neurodevelopment and myelin
Aloysius Domingo, Rachita Yadav, Shivangi Shah, William T. Hendriks, Serkan Erdin, Dadi Gao, Kathryn O’Keefe, Benjamin Currall, James F. Gusella, Nutan Sharma, Laurie J. Ozelius, Michelle E. Ehrlich, Michael E. Talkowski, D. Cristopher Bragg
Inhibition of N-myristoyltransferase Promotes Naive Pluripotency in Mouse and Human Pluripotent Stem Cells
Junko Yoshida, Hitomi Watanabe, Kaori Yamauchi, Takumi Nishikubo, Ayako Isotani, Satoshi Ohtsuka, Hitoshi Niwa, Hidenori Akutsu, Akihiro Umezawa, Hirofumi Suemori, Yasuhiro Takashima, Gen Kondoh, Junji Takeda, Kyoji Horie
The extracellular matrix controls stem cell specification and crypt morphology in the developing and adult gut
R. Ramadan, SM. van Neerven, VM. Wouters, T. Martins Garcia, V. Muncan, OD. Franklin, M. Battle, KS. Carlson, J. Leach, OJ. Sansom, L. Vermeulen, JP. Medema, DJ. Huels
Stem cell-free therapy for glaucoma to preserve vision
Ajay Kumar, Xiong Siqi, Minwen Zhou, Wen Chen, Enzhi Yang, Andrew Price, Liang Le, Ying Zhang, Laurence Florens, Michael Washburn, Akshay Kumar, Yunshu Li, Yi Xu, Kira Lathrop, Katherine Davoli, Yuanyuan Chen, Joel S. Schuman, Ting Xie, Yiqin Du
Tfap2b specifies an embryonic melanocyte stem cell population that retains adult multi-fate potential
Alessandro Brombin, Daniel J. Simpson, Jana Travnickova, Hannah R. Brunsdon, Zhiqiang Zeng, Yuting Lu, Tamir Chandra, E. Elizabeth Patton
Single-cell RNA sequencing-based characterization of resident lung mesenchymal stromal cells in bronchopulmonary dysplasia
I. Mižíková, F. Lesage, C. Cyr-Depauw, D. P. Cook, M. Hurskainen, S.M. Hänninen, A. Vadivel, P. Bardin, S. Zhong, O. Carpen, B. C. Vanderhyden, B. Thébaud
Human iPSC-derived cerebral organoids model features of Leigh Syndrome and reveal abnormal corticogenesis
Alejandra I. Romero-Morales, Gabriella L. Robertson, Anuj Rastogi, Megan L. Rasmussen, Hoor Temuri, Gregory Scott McElroy, Ram Prosad Chakrabarty, Lawrence Hsu, Paula M. Almonacid, Bryan A. Millis, Navdeep S. Chandel, Jean-Philippe Cartailler, Vivian Gama
Zebrafish pigment cells develop directly from persistent highly multipotent progenitors
Masataka Nikaido, Tatiana Subkhankulova, Leonid A. Uroshlev, Artem J. Kasianov, Karen Camargo Sosa, Gemma Bavister, Xueyan Yang, Frederico S. L. M. Rodrigues, Thomas J. Carney, Hartmut Schwetlick, Jonathan H.P. Dawes, Andrea Rocco, Vsevelod Makeev, Robert N. Kelsh
Basal neural stem cells drive postnatal neurogenesis whereas apical stem cells act as proliferation gatekeepers by regulating notch activation in the postnatal ventricular-subventricular zone
Katja Baur, Yomn Abdullah, Claudia Mandl, Gabriele Hoelzl-Wenig, Yan Shi, Udo Schmidt-Edelkraut, Priti Khatri, Francesca Ciccolini
The role of Kabuki Syndrome genes KMT2D and KDM6A in development: Analysis in Human sequencing data and compared to mice and zebrafish
Rwik Sen, Ezra Lencer, Elizabeth A. Geiger, Kenneth Jones, Tamim H. Shaikh, Kristin Bruk Artinger
Single-cell transcriptome analysis of embryonic and adult endothelial cells allows to rank the hemogenic potential of post-natal endothelium
Artem Adamov, Yasmin Natalia Serina Secanechia, Christophe Lancrin
Control of Arabidopsis shoot stem cell homeostasis by two antagonistic CLE peptide signalling pathways
Jenia Schlegel, Grégoire Denay, Karine Gustavo Pinto, Yvonne Stahl, Julia Schmid, Patrick Blümke, Rüdiger Simon
Skeletal dysplasia-causing TRPV4 mutations suppress the hypertrophic differentiation of human iPSC-derived chondrocytes
Amanda R. Dicks, Grigory I. Maksaev, Zainab Harissa, Alireza Savadipour, Ruhang Tang, Nancy Steward, Wolfgang Liedtke, Colin G. Nichols, Chia-Lung Wu, Farshid Guilak
ZFP541 is indispensable for pachytene progression by interacting with KCTD19 and activates meiotic gene expression in mouse spermatogenesis
Yushan Li, Ranran Meng, Shanze Li, Bowen Gu, Xiaotong Xu, Haihang Zhang, Tianyu Shao, Jiawen Wang, Yinghua Zhuang, Fengchao Wang
Generation of liver organoids from human induced pluripotent stem cells as liver fibrosis and steatosis models
Hoi Ying Tsang, Paulisally Hau Yi Lo, Kenneth Ka Ho Lee
Oncofetal protein CRIPTO regulates wound healing and fibrogenesis in regenerating liver and is associated with the initial stages of cardiac fibrosis
Sofia Karkampouna, Danny van der Helm, Bart van Hoek, Hein W Verspaget, Marie Jose TH Goumans, Minneke Coenraad, Boudewijn TH Kruithof, Marianna Kruithof-deJulio
Unbiased in vivo exploration of nuclear bodies-enhanced sumoylation reveals that PML orchestrates embryonic stem cell fate
Sarah Tessier, Omar Ferhi, Marie-Claude Geoffroy, Roman Gonzalez-Prieto, Antoine Canat, Samuel Quentin, Marika Pla, Michiko Niwa-Kawakita, Pierre Bercier, Domitille Rerolle, Pierre Therizols, Emmanuelle Fabre, Alfred C.O. Vertegaal, Hugues de The, Valerie Lallemand-Breitenbach
Distinct epicardial gene regulatory programmes drive development and regeneration of the zebrafish heart
Michael Weinberger, Filipa C. Simoes, Tatjana Sauka-Spengler, Paul R. Riley
Kidney organoids: A system to study human basement membrane assembly in health and disease
Mychel RPT Morais, Pinyuan Tian, Craig Lawless, Syed Murtuza-Baker, Louise Hopkinson, Steven Woods, Aleksandr Mironov, David A Long, Daniel Gale, Telma MT Zorn, Roy Zent, Rachel Lennon
Snail maintains the stem/progenitor state of skin epithelial cells and carcinomas through the autocrine effect of the matricellular protein Mindin
Krithika Badarinath, Binita Dam, Sunny Kataria, Ravindra K. Zirmire, Rakesh Dey, Randhir Singh, Tafheem A. Masudi, Janani Sambath, Prashanth Kumar, Akash Gulyani, You-Wen He, Sudhir Krishna, Colin Jamora
Glypican-6 deficiency causes dose-dependent conotruncal congenital heart malformations through abnormal remodelling of the endocardial cushions
Gennadiy Tenin, Alexander Crozier, Kathryn E. Hentges, Bernard Keavney
TLR4 regulation in human fetal membranes as an explicative mechanism of a pathological preterm case
Corinne Belville, Flora Ponelle-Chachuat, Marion Rouzaire, Christelle Gross, Bruno Pereira, Denis Gallot, Vincent Sapin, Loïc Blanchon
Cell-autonomous differentiation of human primed embryonic stem cells into trophoblastic syncytia through the nascent amnion-like cell state
Masatoshi Ohgushi, Mototsugu Eiraku
Intrinsic and extrinsic regulation of human fetal bone marrow haematopoiesis and perturbations in Down syndrome
Laura Jardine, Simone Webb, Issac Goh, Mariana Quiroga Londoño, Gary Reynolds, Michael Mather, Bayanne Olabi, Emily Stephenson, Rachel A. Botting, Dave Horsfall, Justin Engelbert, Daniel Maunder, Nicole Mende, Caitlin Murnane, Emma Dann, Jim McGrath, Hamish King, Iwo Kucinski, Rachel Queen, Christopher D Carey, Caroline Shrubsole, Elizabeth Poyner, Meghan Acres, Claire Jones, Thomas Ness, Rowan Coulthard, Natalina Elliott, Sorcha O’Byrne, Myriam L. R. Haltalli, John E Lawrence, Steven Lisgo, Petra Balogh, Kerstin B Meyer, Elena Prigmore, Kirsty Ambridge, Mika Sarkin Jain, Mirjana Efremova, Keir Pickard, Thomas Creasey, Jaume Bacardit, Deborah Henderson, Jonathan Coxhead, Andrew Filby, Rafiqul Hussain, David Dixon, David McDonald, Dorin-Mirel Popescu, Monika S. Kowalczyk, Bo Li, Orr Ashenberg, Marcin Tabaka, Danielle Dionne, Timothy L. Tickle, Michal Slyper, Orit Rozenblatt-Rosen, Aviv Regev, Sam Behjati, Elisa Laurenti, Nicola K. Wilson, Anindita Roy, Berthold Göttgens, Irene Roberts, Sarah A. Teichmann, Muzlifah Haniffa
Inflammatory blockade prevents injury to the developing pulmonary gas exchange surface in preterm primates
Andrea Toth, Shelby Steinmeyer, Paranthaman Kannan, Jerilyn Gray, Courtney M. Jackson, Shibabrata Mukherjee, Martin Demmert, Joshua R. Sheak, Daniel Benson, Joe Kitzmiller, Joseph A. Wayman, Pietro Presicce, Christopher Cates, Rhea Rubin, Kashish Chetal, Yina Du, Yifei Miao, Mingxia Gu, Minzhe Guo, Vladimir V. Kalinichenko, Suhas G. Kallapur, Emily R. Miraldi, Yan Xu, Daniel Swarr, Ian Lewkowich, Nathan Salomonis, Lisa Miller, Jennifer S. Sucre, Jeffrey A. Whitsett, Claire A. Chougnet, Alan H. Jobe, Hitesh Deshmukh, William J. Zacharias
Post-embryonic development and aging of the appendicular skeleton in Ambystoma mexicanum
Camilo Riquelme-Guzmán, Maritta Schuez, Alexander Böhm, Dunja Knapp, Sandra Edwards-Jorquera, Alberto S. Ceccarelli, Osvaldo Chara, Martina Rauner, Tatiana Sandoval-Guzmán
| Plant development
Populus ERF85 balances xylem cell expansion and secondary cell wall formation in hybrid aspen
Carolin Seyfferth, Bernard A Wessels, Jorma Vahala, Jaakko Kangasjarvi, Nicolas Delhomme, Torgeir R Hvidsten, Hannele Tuominen, Judith Felten
B1L regulates lateral root development by exocytic vesicular trafficking-mediated polar auxin transport in Arabidopsis
Gang Yang, Bi-xia Chen, Tao Chen, Jia-hui Chen, Rui Sun, Cong-cong Liu, Jiao Jia, Xiu-le Yue, Li-zhe An, Hua Zhang
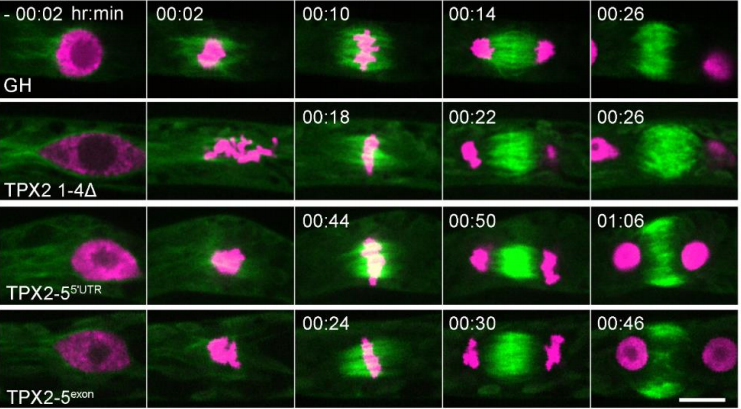
Spindle position dictates division site during asymmetric cell division in moss
Elena Kozgunova, Mari W. Yoshida, Ralf Reski, Gohta Goshima
Maize Brittle Stalk2-Like3, encoding a COBRA protein, functions in cell wall formation and carbohydrate partitioning
Benjamin T. Julius, Tyler J. McCubbin, Rachel A. Mertz, Nick Baert, Jan Knoblauch, DeAna G. Grant, Kyle Conner, Saadia Bihmidine, Paul Chomet, Ruth Wagner, Jeff Woessner, Karen Grote, Jeanette Peevers, Thomas L. Slewinski, Maureen C. McCann, Nicholas C. Carpita, Michael Knoblauch, David M. Braun
Protein turnover in the developing Triticum aestivum grain
Hui Cao, Owen Duncan, A. Harvey Millar
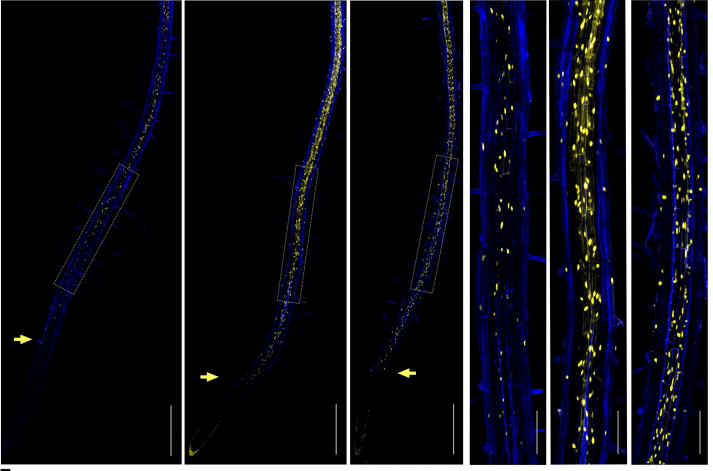
Suberin plasticity to developmental and exogenous cues is regulated by a set of MYB transcription factors
Vinay Shukla, Jian-Pu Han, Fabienne Cléard, Linnka Lefebvre- Legendre, Kay Gully, Paulina Flis, Alice Berhin, Tonni Grube Andersen, David E Salt, Christiane Nawrath, Marie Barberon
Endogenous RNA editing of a nuclear gene BOSS triggers flowering in tomato
Wenqian Wang, Jie Ye, Chuying Yu, Qingmin Xie, Xin Wang, Huiyang Yu, Jianwen Song, Changxing Li, Long Cui, Heyou Han, Changxian Yang, Hanxia Li, Yongen Lu, Taotao Wang, Yuyang Zhang, Junhong Zhang, Bo Ouyang, Zhibiao Ye
Transcriptomic analysis of temporal shifts in berry development between two grapevine cultivars of the Pinot family reveals potential genes controlling ripening time
Jens Theine, Daniela Holtgräwe, Katja Herzog, Florian Schwander, Anna Kicherer, Ludger Hausmann, Prisca Viehöver, Reinhard Töpfer, Bernd Weisshaar
Chromatin enrichment for Proteomics in Plants (ChEP-P) implicates the histone reader ALFIN-LIKE 6 in jasmonate signalling
Isabel Cristina Vélez-Bermúdez, Wolfgang Schmidt
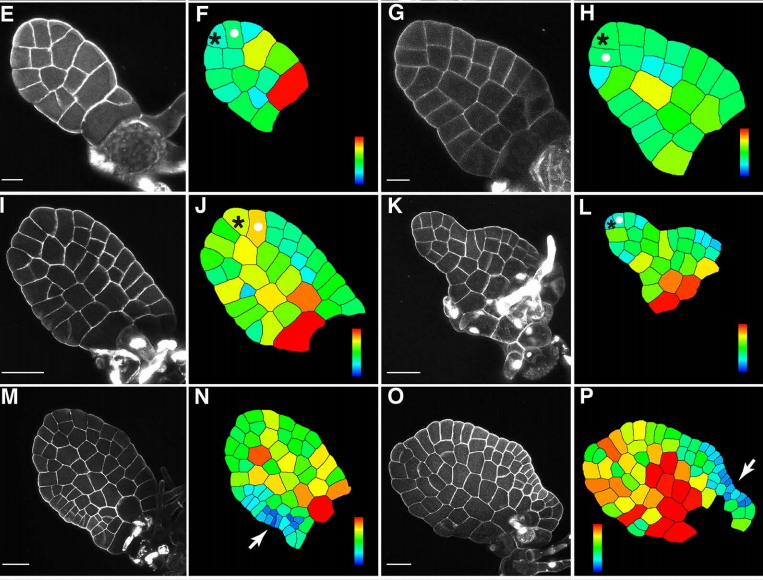
Timing of meristem initiation and maintenance determines the morphology of fern gametophytes
Xiao Wu, An Yan, Scott McAdam, Jo Ann Banks, Shaoling Zhang, Yun Zhou
The Genetic Architecture of Strawberry Yield and Fruit Quality Traits
Helen M. Cockerton, Amanda Karlström, Abigail W. Johnson, Bo Li, Eleftheria Stavridou, Katie J. Hopson, Adam B. Whitehouse, Richard J. Harrison
The REF6-dependent H3K27 demethylation establishes transcriptional competence to promote germination in Arabidopsis
Jie Pan, Huairen Zhang, Zhenping Zhan, Ting Zhao, Danhua Jiang
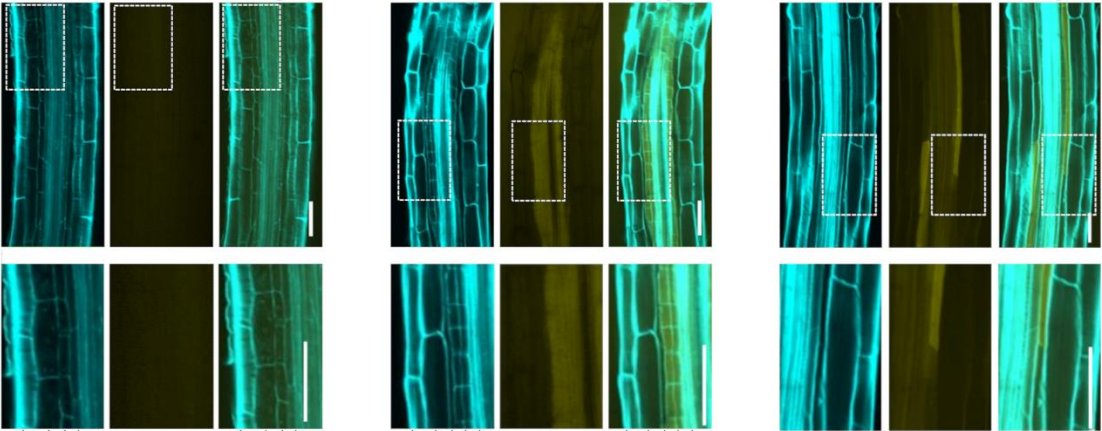
Regulation of suberin biosynthesis and Casparian strip development in the root endodermis by two plant auxins
Sam David Cook, Seisuke Kimura, Qi Wu, Rochus Benni Franke, Takehiro Kamiya, Hiroyuki Kasahara
Estimation of cell cycle kinetics in higher plant root meristem links organ position with cellular fate and chromatin structure
Taras Pasternak, Stefan Kircher, Klaus Palme
The VIL gene CRAWLING ELEPHANT controls maturation and differentiation in tomato via polycomb silencing
Ido Shwartz, Chen Yahav, Neta Kovetz, Alon Israeli, Maya Bar, Matan Levy, Katherine L. Duval, José M. Jiménez-Gómez, Roger B. Deal, Naomi Ori
A quantitative gibberellin signalling biosensor reveals a role for gibberellins in internode specification at the shoot apical meristem
Bihai Shi, Amelia Felipo-Benavent, Guillaume Cerutti, Carlos Galvan-Ampudia, Lucas Jilli, Geraldine Brunoud, Jérome Mutterer, Lali Sakvarelidze-Achard, Jean-Michel Davière, Alejandro Navarro-Galiano, Ankit Walia, Shani Lazary, Jonathan Legrand, Roy Weinstein, Alexander M. Jones, Salomé Prat, Patrick Achard, Teva Vernoux
Adaptive reprogramming during early seed germination requires temporarily enhanced fermentation – a critical role for alternative oxidase (AOX) regulation that concerns also microbiota effectiveness
Bharadwaj Revuru, Carlos Noceda, Mohanapriya Gunasekaran, Sarma Rajeev Kumar, Karine Leitão Lima Thiers, José Hélio Costa, Elisete Santos Macedo, Aprajita Kumari, Kapuganti Jagadis Gupta, Shivani Srivastava, Alok Adholeya, Manuela Oliveira, Isabel Velada, Debabrata Sircar, Ramalingam Sathishkumar, Birgit Arnholdt-Schmitt
Ovule siRNAs methylate and silence protein-coding genes in trans
Diane Burgess, Hiu Tung Chow, Jeffrey W. Grover, Michael Freeling, Rebecca A. Mosher
SlKIX8 and SlKIX9 are negative regulators of leaf and fruit growth in tomato
Gwen Swinnen, Jean-Philippe Mauxion, Alexandra Baekelandt, Rebecca De Clercq, Jan Van Doorsselaere, Dirk Inzé, Nathalie Gonzalez, Alain Goossens, Laurens Pauwels
Shade-induced WRKY transcription factors restrict root growth during the shade avoidance response
Daniele Rosado, Amanda Ackermann, Olya Spassibojko, Magdalena Rossi, Ullas V. Pedmale
The U1 snRNP component RBP45d regulates temperature-responsive flowering in Arabidopsis thaliana
Ping Chang, Hsin-Yu Hsieh, Shih-Long Tu
INDEHISCENT regulates explosive seed dispersal
Anahit Galstyan, Penny Sarchet, Rafael Campos-Martin, Milad Adibi, Lachezar A. Nikolov, Miguel Pérez Antón, Léa Rambaud-Lavigne, Xiangchao Gan, Angela Hay
Seed morphological traits as a tool to quantify variation maintained in ex situ collections: a case study in Pinus torreyana (Parry)
Lionel N Di Santo, Monica Polgar, Storm Nies, Paul Hodgkiss, Courtney A Canning, Jessica W Wright, Jill A Hamilton
DNA METHYLTRANSFERASE 3 (MET3) is regulated by Polycomb Group complex during Arabidopsis endosperm development
Louis Tirot, Pauline E. Jullien
Developmental Effects on Relative Use of PEPCK and NADP-ME Pathways of C4 Photosynthesis in Maize
Jennifer J. Arp, Shrikaar Kambhampati, Kevin L. Chu, Somnath Koley, Lauren M. Jenkins, Todd C. Mockler, Doug K. Allen
GTL1 is required for a robust root hair growth response to avoid nutrient overloading
Michitaro Shibata, David S. Favero, Ryu Takebayashi, Ayako Kawamura, Bart Rymen, Yoichiroh Hosokawa, Keiko Sugimoto
Transcriptional activation of auxin biosynthesis drives developmental reprogramming of differentiated cells
Yuki Sakamoto, Ayako Kawamura, Takamasa Suzuki, Shoji Segami, Masayoshi Maeshima, Stefanie Polyn, Lieven De Veylder, Keiko Sugimoto
Genetic basis and dual adaptive role of floral pigmentation in sunflowers
Marco Todesco, Natalia Bercovich, Amy Kim, Ivana Imerovski, Gregory L. Owens, Óscar Dorado Ruiz, Srinidhi V. Holalu, Lufiani L. Madilao, Mojtaba Jahani, Jean-Sébastien Légaré, Benjamin K. Blackman, Loren H. Rieseberg
Arabidopsis stomatal polarity protein BASL mediates distinct processes before and after cell division to coordinate cell size and fate asymmetries
Yan Gong, Julien Alassimone, Andrew Muroyama, Gabriel Amador, Rachel Varnau, Ao Liu, Dominique C. Bergmann
Arabidopsis ABIG1 Functions in Laminar Growth and Polarity Formation through Regulation by REVOLUTA and KANADI
Jesus Preciado, Kevin Begcy, Tie Liu
3D reconstruction identifies loci linked to variation in angle of individual sorghum leaves
Michael C. Tross, Mathieu Gaillard, Mackenzie Zweiner, Chenyong Miao, Bosheng Li, Bedrich Benes, James C. Schnable
Members of the ELMOD protein family specify formation of distinct aperture domains on the Arabidopsis pollen surface
Yuan Zhou, Prativa Amom, Sarah H. Reeder, Byung Ha Lee, Adam Helton, Anna A. Dobritsa
Ureides are similarly accumulated in response to UV-C irradiation and wound but differently remobilized during recovery in Arabidopsis leaves.
Aigerim Soltabayeva, Aizat Bekturova, Assylay Kurmanbayeva, Dinara Oshanova, Zhadyrassyn Nurbekova, Sudhakar Srivastava, Dominic Standing, Moshe Sagi
| Evo-devo
Evolution of the nitric oxide synthase family in vertebrates and novel insights in gill development
Giovanni Annona, Iori Sato, Juan Pascual-Anaya, Ingo Braasch, Randal Voss, Jan Stundl, Vladimir Soukup, Sihigeru Kuratani, John Postlethwait, Salvatore D’Aniello
Evolutionary dynamics of sex-biased genes expressed in cricket brains and gonads
Carrie A. Whittle, Arpita Kulkarni, Cassandra G. Extavour
Single-nucleus transcriptomes reveal functional and evolutionary properties of cell types in the Drosophila accessory gland
Alex C. Majane, Julie M. Cridland, David J. Begun
Evolutionary transition of doublesex regulation in termites and cockroaches: from sex-specific splicing to male-specific transcription
Satoshi Miyazaki, Kokuto Fujiwara, Keima Kai, Yudai Masuoka, Hiroki Gotoh, Teruyuki Niimi, Yoshinobu Hayashi, Shuji Shigenobu, Kiyoto Maekawa
Complete metamorphosis and microbiota turnover in insects
Christin Manthey, Paul R Johsnton, Jens Rolff
Facultative release from developmental constraints through polyphenism promotes adaptively flexible maturation
Flor T. Rhebergen, Isabel M. Smallegange
A large disordered region confers a wide spanning volume to vertebrate Suppressor of Fused as shown in a trans-species solution study
Staëlle Makamte, Amira Jabrani, Annick Paquelin, Anne Plessis, Mathieu Sanial, Aurélien Thureau, Olga Rudenko, Francesco Oteri, Marc Baaden, Valérie Biou
Developmental plasticity in male courtship in Bicyclus anynana butterflies is driven by hormone regulation of the yellow gene
Heidi Connahs, Eunice Jingmei Tan, Yi Ting Ter, Emilie Dion, Yuji Matsuoka, Ashley Bear, Antónia Monteiro
Convergent adaptation and ecological speciation result from unique genomic mechanisms in sympatric extremophile fishes
Ryan Greenway, Anthony P. Brown, Henry Camarillo, Cassandra Delich, Kerry L. McGowan, Joel Nelson, Lenin Arias-Rodriguez, Joanna L. Kelley, Michael Tobler
Behavioural adaptations in egg laying ancestors facilitate evolutionary transitions to live birth
Amanda K. Pettersen, Nathalie Feiner, Daniel W.A. Noble, Geoffrey M. While, Charlie K. Cornwallis, Tobias Uller
Functional divergence of the bag of marbles gene in the Drosophila melanogaster species group
Jaclyn E. Bubnell, Cynthia K.S. Ulbing, Paula Fernandez-Begne, Charles F. Aquadro
Adaptive shifts underlie the divergence in wing morphology in bombycoid moths
Brett R. Aiello, Milton Tan, Usama Bin Sikandar, Alexis J. Alvey, Burhanuddin Bhinderwala, Katalina C. Kimball, Jesse R. Barber, Chris A. Hamilton, Akito Y. Kawahara, Simon Sponberg
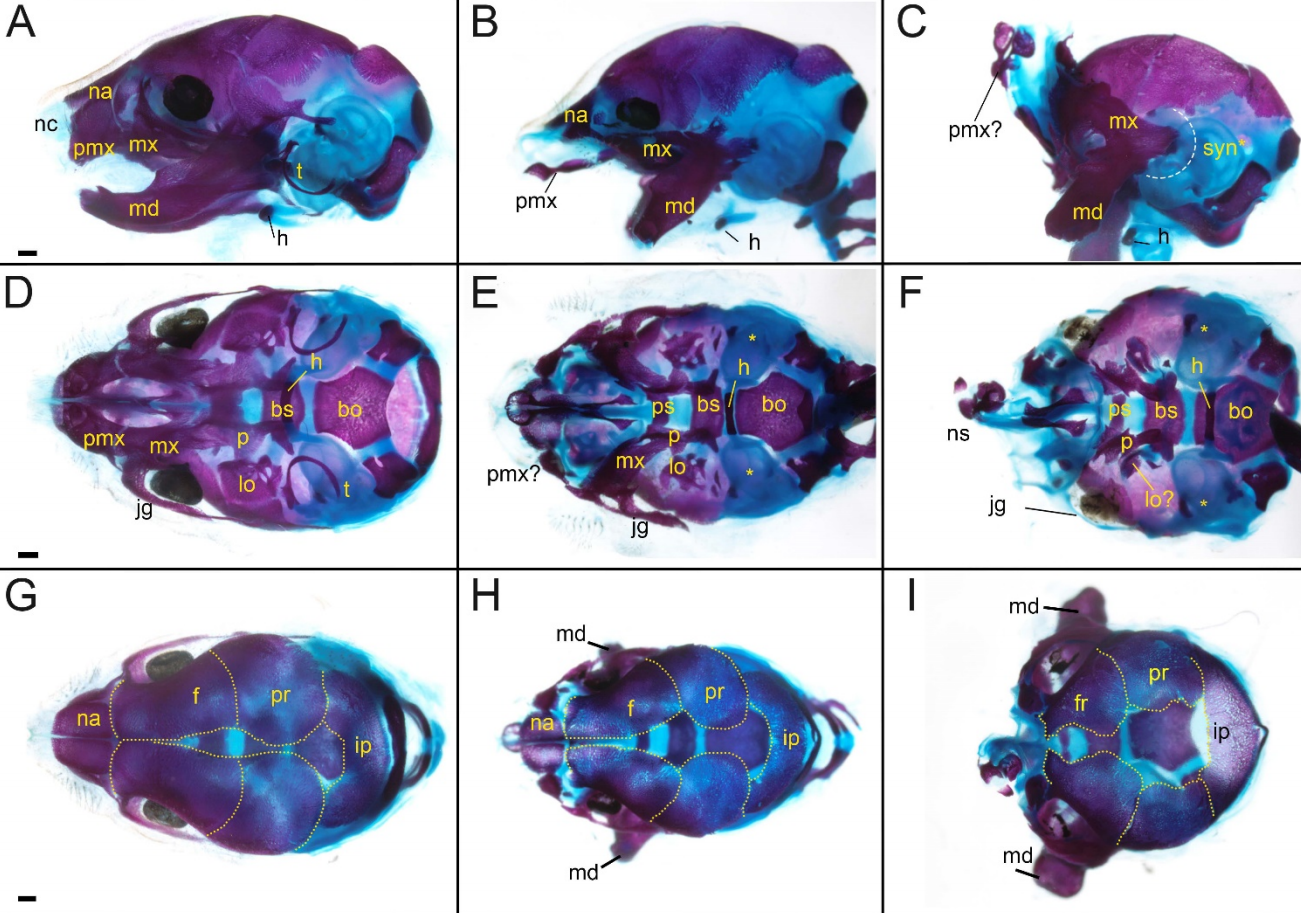
An evolutionarily conserved odontode gene regulatory network underlies head armor formation in suckermouth armored catfish
Shunsuke Mori, Tetsuya Nakamura
Evolution of lbx spinal cord expression and function
José Luis Juárez-Morales, Frida Weierud, Samantha J. England, Celia Demby, Nicole Santos, Ginny Grieb, Sylvie Mazan, Katharine E. Lewis
Evolution of Drosophila buzzatii wings: Modular genetic organization, sex-biased integrative selection and intralocus sexual conflict
PP Iglesias, FA Machado, S Llanes, E Hasson, EM Soto
Sex-Specific Plasticity Explains Genetic Variation in Sexual Size Dimorphism in Drosophila
Isabelle M Vea, Austin Wilcox, W. Anthony Frankino, Alexander W Shingleton
Cell Biology
A mechano-osmotic feedback couples cell volume to the rate of cell deformation
Larisa Venkova, Amit Singh Vishen, Sergio Lembo, Nishit Srivastava, Baptiste Duchamp, Artur Ruppel, Stéphane Vassilopoulos, Alexandre Deslys, Juan Manuel Garcia Arcos, Alba Diz-Muñoz, Martial Balland, Jean-François Joanny, Damien Cuvelier, Pierre Sens, Matthieu Piel
Volume growth in animal cells is cell cycle dependent and shows additive fluctuations
Clotilde Cadart, Matthieu Piel, Marco Cosentino Lagomarsino
A particle size threshold governs diffusion and segregation of PAR-3 during cell polarization
Yiran Chang, Daniel J. Dickinson
KIF18B is a cell-type specific regulator of spindle orientation in the epidermis
Rebecca S. Moreci, Terry Lechler
A ciliopathy complex builds distal appendages to initiate ciliogenesis
Dhivya Kumar, Addison Rains, Vicente Herranz-Pérez, Quanlong Lu, Xiaoyu Shi, Danielle L. Swaney, Erica Stevenson, Nevan J. Krogan, Bo Huang, Christopher Westlake, Jose Manuel Garcia-Verdugo, Bradley Yoder, Jeremy F. Reiter
Mouse oocytes do not contain a Balbiani body
Laasya Dhandapani, Marion C. Salzer, Juan M. Duran, Gabriele Zaffagnini, Cristian De Guirior, Maria Angeles Martínez-Zamora, Elvan Böke
SLC1A5 provides glutamine and asparagine necessary for bone development in mice
Deepika Sharma, Yilin Yu, Leyao Shen, Guo-Fang Zhang, Courtney Karner
The Transcriptional Co-Activator Taz Contributes to the Differentiation of a Salivary Gland Epithelial Cell Line Towards a Myoepithelial Phenotype
Renee F. Thiemann, Scott Varney, Nicholas Moskwa, John Lamar, Melinda Larsen, Susan E. LaFlamme
Anoikis resistance in mammary epithelial cells is mediated by semaphorin 7a
Taylor R. Rutherford, Alan M Elder, Traci R. Lyons
A steroid hormone regulates growth in response to oxygen availability
George P. Kapali, Viviane Callier, Hailey Broeker, Parth Tank, Samuel J.L. Gascoigne, Jon F Harrison, Alexander W. Shingleton
Modelling
Cell types and ontologies of the Human Cell Atlas
David Osumi-Sutherland, Chuan Xu, Maria Keays, Peter V. Kharchenko, Aviv Regev, Ed Lein, Sarah A. Teichmann
Computational modelling of cell motility modes emerging from cell-matrix adhesion dynamics
Leonie van Steijn, Clément Sire, Loïc Dupré, Guy Theraulaz, Roeland M.H. Merks
A landscape model for cell fate decisions during mesoendoderm differentiation in C. elegans based on Wnt dynamics
Shyr-Shea Chang, Zhirong Bao, Eric D. Siggia
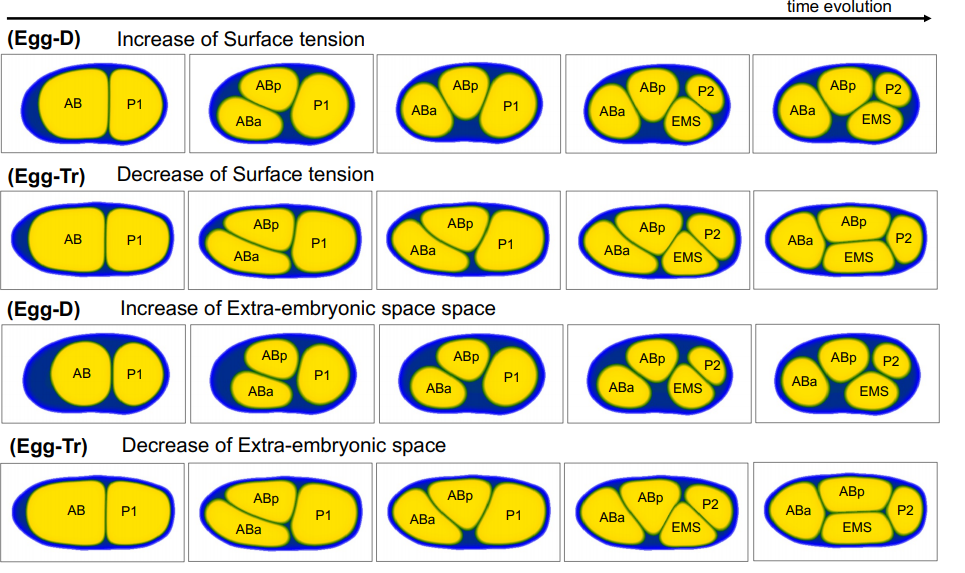
The extra-embryonic space is a geometric constraint regulating cell arrangement in nematodes
Sungrim Seirin-Lee, Akatsuki Kimura
From heterogeneous datasets to predictive models of embryonic development
Sayantan Dutta, Aleena L. Patel, Shannon E. Keenan, Stanislav Y. Shvartsman
Graph-based machine learning reveals rules of spatiotemporal cell interactions in tissues
Takaki Yamamoto, Katie Cockburn, Valentina Greco, Kyogo Kawaguchi
Blastocoel morphogenesis: a biophysics perspective
Mathieu Le-Verge-Serandour, Hervé Turlier
Disorder in cellular packing can alter proliferation dynamics to regulate growth
Chandrashekar Kuyyamudi, Shakti N. Menon, Fernando Casares, Sitabhra Sinha
Tools & Resources
hei-tag: a highly efficient tag to boost targeted genome editing
Thomas Thumberger, Tinatini Tavhelidse, Jose Arturo Gutierrez-Triana, Rebekka Medert, Alex Cornean, Bettina Welz, Marc Freichel, Joachim Wittbrodt
NanoDam identifies novel temporal transcription factors conserved between the Drosophila central brain and visual system
Jocelyn L.Y. Tang, Anna E. Hakes, Robert Krautz, Takumi Suzuki, Esteban G. Contreras, Paul M. Fox, Andrea H. Brand
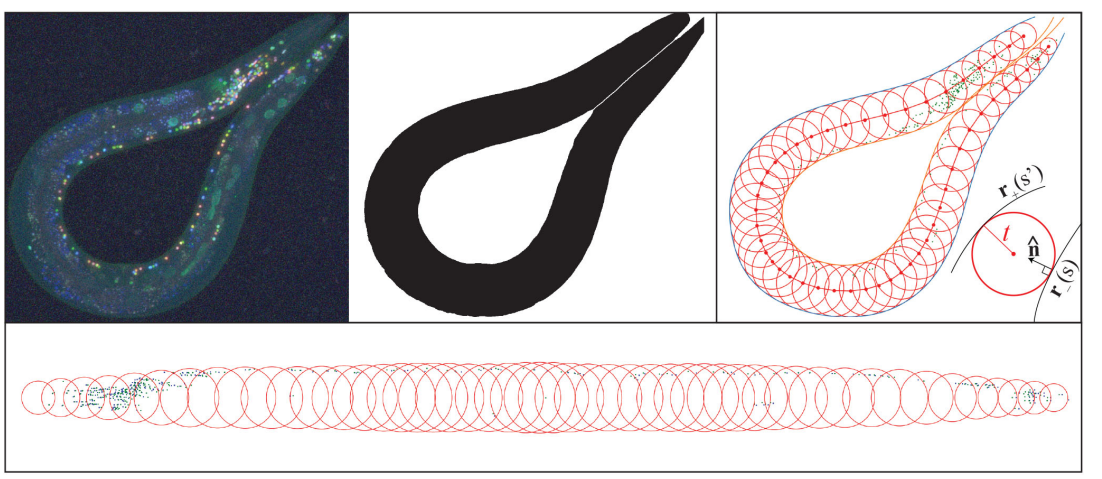
Toward a More Accurate 3D Atlas of C. elegans Neurons
Michael Skuhersky, Tailin Wu, Eviatar Yemini, Edward Boyden, Max Tegmark
Genome editing in animals with minimal PAM CRISPR-Cas9 enzymes
Jeremy Vicencio, Carlos Sánchez-Bolaños, Ismael Moreno-Sánchez, David Brena, Dmytro Kukhtar, Miguel Ruiz-López, Mariona Cots-Ponjoan, Charles E. Vejnar, Alejandro Rubio, Natalia Rodrigo Melero, Carlo Carolis, Antonio J. Pérez-Pulido, Antonio J. Giráldez, Benjamin P. Kleinstiver, Julián Cerón, Miguel A. Moreno-Mateos
A CRISPR toolbox for generating intersectional genetic mice for functional, molecular, and anatomical circuit mapping
Savannah J. Lusk, Andrew McKinney, Patrick J. Hunt, Paul G. Fahey, Jay Patel, Jenny J. Sun, Vena K. Martinez, Ping Jun Zhu, Jeremy R. Egbert, Xiaolong Jiang, Benjamin R. Arenkiel, Andreas S. Tolias, Mauro Costa-Mattioli, Russell S. Ray
Extremely bright, near-IR emitting spontaneously blinking fluorophores enable ratiometric multicolor nanoscopy in live cells
Jonathan Tyson, Kevin Hu, Shuai Zheng, Phylicia Kidd, Neville Dadina, Ling Chu, Derek Toomre, Joerg Bewersdorf, Alanna Schepartz
CeLINC, a fluorescence-based protein-protein interaction assay in C. elegans
Jason R Kroll, Sanne Remmelzwaal, Mike Boxem
Anatomical Structures, Cell Types, and Biomarkers Tables Plus 3D Reference Organs in Support of a Human Reference Atlas
Katy Börner, Sarah A. Teichmann, Ellen M. Quardokus, James Gee, Kristen Browne, David Osumi-Sutherland, Bruce W. Herr II, Andreas Bueckle, Hrishikesh Paul, Muzlifah A. Haniffa, Laura Jardine, Amy Bernard, Song-Lin Ding, Jeremy A. Miller, Shin Lin, Marc Halushka, Avinash Boppana, Teri A. Longacre, John Hickey, Yiing Lin, M. Todd Valerius, Yongqun He, Gloria Pryhuber, Xin Sun, Marda Jorgensen, Andrea J. Radtke, Clive Wasserfall, Fiona Ginty, Jonhan Ho, Joel Sunshine, Rebecca T. Beuschel, Maigan Brusko, Sujin Lee, Rajeev Malhotra, Sanjay Jain, Griffin Weber
Fast 3D Clear: A Fast, Aqueous-Based, Reversible Three-Day Tissue Clearing Method for Adult and Embryonic Mouse Brain and Whole Body
Stylianos Kosmidis, Adrian Negrean, Alex Dranovsky, Attila Losonczy, Eric R. Kandel
The developmental transcriptome of Parhyale hawaiensis: microRNAs and mRNAs show different expression dynamics during the maternal-zygotic transition
Llilians Calvo, Maria Birgaoanu, Tom Pettini, Matthew Ronshaugen, Sam Griffiths-Jones
Tissue-specific modification of cellular bioelectrical activities using the chemogenetic tool, DREADD, in zebrafish
Martin R. Silic, GuangJun Zhang
Single nucleus pituitary transcriptomic and epigenetic landscape reveals human stem cell heterogeneity with diverse regulatory mechanisms
Zidong Zhang, Michel Zamojski, Gregory R. Smith, Thea L. Willis, Val Yianni, Natalia Mendelev, Hanna Pincas, Nitish Seenarine, Mary Anne S. Amper, Mital Vasoya, Venugopalan D. Nair, Judith L. Turgeon, Daniel J. Bernard, Olga G. Troyanskaya, Cynthia L. Andoniadou, Stuart C. Sealfon, Frederique Ruf-Zamojski
Zebrafish Cre/lox regulated UFlip alleles generated by CRISPR/Cas targeted integration provide cell-type specific conditional gene inactivation
Maira P. Almeida, Sekhar Kambakam, Fang Liu, Zhitao Ming, Jordan M. Welker, Wesley A. Wierson, Laura E. Schultz-Rogers, Stephen C. Ekker, Karl J. Clark, Jeffrey J. Essner, Maura McGrail
A highly efficient reporter system for identifying and characterizing in vitro expanded hematopoietic stem cells
James L.C. Che, Daniel Bode, Iwo Kucinski, Alyssa H. Cull, Fiona Bain, Melania Barile, Grace Boyd, Miriam Belmonte, Maria Jassinskaja, Juan Rubio-Lara, Mairi S. Shepherd, Anna Clay, Adam C. Wilkinson, Hiromitsu Nakauchi, Satoshi Yamazaki, Berthold Göttgens, David G. Kent
Forward genetics combined with unsupervised classifications identified zebrafish mutants affecting biliary system formation
Divya Jyoti Singh, Kathryn M. Tuscano, Karen L. Ortega, Manali Dimri, Kevin Tae, William Lee, Muslim A. Muslim, Jay L. Liu, Lain X. Pierce, Allyson McClendon, Gregory Naegele, Isabel Gibson, Jodi Livesay, Takuya F. Sakaguchi
Research practice & education
Building Back More Equitable STEM Education: Teach Science by Engaging Students in Doing Science
Sarah C R Elgin, Shan Hays, Vida Mingo, Christopher D Shaffer, Jason Williams
TeamTree analysis: a new approach to evaluate scientific production
Frank W. Pfrieger
I, We, and They: A Linguistic and Narrative Exploration of the Authorship Process
Abigail Konopasky, Bridget C O’Brien, Anthony R Artino Jr., Erik W Driessen, Christopher J Watling, Lauren A Maggio


 (No Ratings Yet)
(No Ratings Yet)