Closing Date: 15 March 2021
A great opportunity for computationally inclined developmental biologists. Note tight deadline.
-D Parichy
The University of Virginia invites applications for a tenure-track Assistant Professor position with joint appointment in the Department of Biology and the School of Data Science. We seek applicants whose research programs address fundamental questions at the interface of Biology and Data Science. Of particular interest are researchers aiming to develop innovative computational tools to improve biological understanding in areas potentially including but not limited to: genomics and phenotype prediction; cell state and signaling; biological network architecture and information processing; multiscale modelling; cellular, organismal or population dynamics; biological image acquisition and analysis. Applicants are sought whose work will synergize with existing labs in the Department of Biology and elsewhere, with research emphases ranging from molecules to cells and tissues, and organisms to populations and ecosystems, as well as programs in the new School for Data Science in the areas of data acquisition, engineering, analysis, visualization or dissemination. Applicants employing computational methods with or without experimental approaches will be considered.
A successful candidate is expected to establish a vigorous, independent, and externally funded research program as well as provide instruction and scientific training at the undergraduate and graduate levels. Applicants with a respect for diversity and a passion for making a positive impact on the world in a collaborative, open environment are strongly encouraged to apply. The position will begin on August 25, 2020.
Located within the College of Arts and Sciences, the Department of Biology provides an interdisciplinary and collaborative environment for basic research and teaching that spans multiple levels of biological organization. The newly formed School of Data Science, founded with the largest gift in the university’s history, is dedicated to open interdisciplinary research of societal benefit with data science at the core. With the schools of Medicine, Engineering & Applied Sciences, UVA offers a diverse, collegial, interdisciplinary, and collaborative environment.
Applicants must have a Ph.D. in life sciences, computer science, statistics or a related field by the start of their appointment. A successful applicant will also have research accomplishments and plans of outstanding quality and significance at the interface of biology and data science as well as a commitment to excellence in teaching and mentoring. A proven commitment to participate in and further develop a diverse, collegial, interdisciplinary, and collaborative environment needs to be demonstrated.
Please apply online at uva.wd1.myworkdayjobs.com/en-US/UVAJobs/job/Charlottesville-VA/Assistant-Professor-of-Biology-Data-Science_R0010887 and attach a cover letter that succinctly highlights your most significant research accomplishments, experiences, and qualifications; a curriculum vitae; a research statement that describes your vision for your research program at the university (≤ 3 pages); a statement of teaching goals; a diversity statement that describes your past experience working on issues of diversity, equity and inclusion and/or working with diverse populations; and the contact information of three references.
Review of applications will begin November 3, 2019; candidates who apply by then will be given priority consideration, but the position will remain open until filled.
 (No Ratings Yet)
(No Ratings Yet)
 Loading...
Loading...


 (No Ratings Yet)
(No Ratings Yet) “Cnidarian Neural Development” (Fabian Rentzsch) at the University of Bergen/Sars Centre in Bergen, Norway.The group uses the sea anemone Nematostella vectensis to study molecular, cellular and evolutionary aspects of nervous system development (e.g. Richards and Rentzsch, Development
“Cnidarian Neural Development” (Fabian Rentzsch) at the University of Bergen/Sars Centre in Bergen, Norway.The group uses the sea anemone Nematostella vectensis to study molecular, cellular and evolutionary aspects of nervous system development (e.g. Richards and Rentzsch, Development 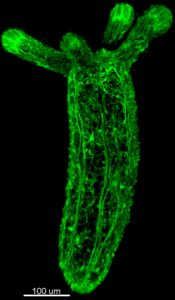

 (1 votes)
(1 votes)