August in preprints
Posted by the Node, on 6 September 2023
Welcome to our monthly trawl for developmental and stem cell biology (and related) preprints.
The preprints this month are hosted on bioRxiv – use these links below to get to the section you want:
- Patterning & signalling
- Morphogenesis & mechanics
- Genes & genomes
- Stem cells, regeneration & disease modelling
- Plant development
- Evo-devo
Developmental biology
| Patterning & signalling
Danielle C. Spitzer, William Y. Sun, Anthony Rodríguez-Vargas, Iswar K. Hariharan
Carlo Donato Caiaffa, Yogeshwari S. Ambekar, Manmohan Singh, Ying Linda Lin, Bogdan Wlodarczyk, Salavat R. Aglyamov, Giuliano Scarcelli, Kirill V. Larin, Richard Finnell
Andrew Liu, Jessica O’Connell, Farley Wall, Richard W. Carthew
Helena Bugacov, Andrew McMahon, Balint Der, Sunghyun Kim, Nils Olof Lindstrom
Fgf signalling is required for gill slit formation in the skate, Leucoraja erinacea
Jenaid M Rees, Michael A Palmer, J. Andrew Gillis
Anastasiia Lozovska, Ana Nóvoa, Ying-Yi Kuo, Arnon Dias Jurberg, Gabriel G Martins, Anna-Katerina Hadjantonakis, Moises Mallo
Nanami Morooka, Ning Gui, Koji Ando, Keisuke Sako, Moe Fukumoto, Melina Hußmann, Stefan Schulte-Merker, Naoki Mochizuki, Hiroyuki Nakajima
Eduardo D. Gigante, Katarzyna M. Piekarz, Alexandra Gurgis, Leslie Cohen, Florian Razy-Krajka, Sydney Popsuj, Hussan S. Ali, Shruthi Mohana Sundaram, Alberto Stolfi
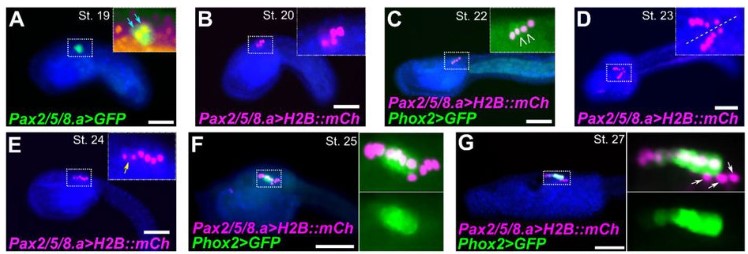
Hedgehog signaling is required for the maintenance of mesenchymal nephron progenitors
Eunah Chung, Patrick Deacon, Yueh-Chiang Hu, Hee-Woong Lim, Joo-Seop Park
Modular control of time and space during vertebrate axis segmentation
Ali Seleit, Ian Brettell, Tomas W Fitzgerald, Carina Vibe, Felix Loosli, Joachim Wittbrodt, Kiyoshi Naruse, Ewan Birney, Alexander Aulehla
Jianbo Wang, Chenbei Chang, Hwa-seon Seo, Deli Yu, Ivan Popov, Jiahui Tao, Allyson Angermeier, Bingdong Sha, Jeffery Axelrod
Epigenetic heterogeneity of the Notch signaling components in the developing human retina
Takahiro Nakayama, Masaharu Yoshihara, Satoru Takahashi
Zn2+ is Essential for Ca2+ Oscillations in Mouse Eggs
Hiroki Akizawa, Emily M Lopes, Rafael A Fissore
Balint Der, Helena Bugacov, Bohdana-Myroslava Briantseva, Andrew P. McMahon
Spatial patterning regulates neuron numbers in the Drosophila visual system
Jennifer Malin, Yen-Chung Chen, Félix Simon, Evelyn Keefer, Claude Desplan
Lianjie Miao, Micah Castillo, Yangyang Lu, Yongqi Xiao, Yu Liu, Alan R Burns, Ashok Kumar, Preethi Gunaratne, C. Michael DiPersio, Mingfu Wu
Spatiotemporal dynamics of cytokines expression dictate fetal liver hematopoiesis
Marcia Mesquita Peixoto, Francisca Soares-da-Silva, Valentin Bonnet, Gustave Ronteix, Rita Faria Santos, Marie-Pierre Mailhe, Xing Feng, João Pedro Pereira, Emanuele Azzoni, Giorgio Anselmi, Marella de Bruijn, Charles N. Baroud, Perpétua Pinto-do-Ó, Ana Cumano
Juan Manuel Gomez, Hendrik Nolte, Elisabeth Vogelsang, Bipasha Dey, Michiko Takeda, Girolamo Giudice, Miriam Faxel, Alina Cepraga, Robert Patrick Zinzen, Marcus Krüger, Evangelia Petsalaki, Yu-Chiun Wang, Maria Leptin
Nicholas X. Russell, Kaulini Burra, Ronak Shah, Natalia Bottasso-Arias, Megha Mohanakrishnan, John Snowball, Harshavardhana H. Ediga, Satish K Madala, Debora Sinner
| Morphogenesis & mechanics
Hierarchical Morphogenesis of Swallowtail Butterfly Wing Scale Nanostructures
Kwi Shan Seah, Vinodkumar Saranathan
Two distinct mechanisms of Plexin A function in Drosophila optic lobe lamination and morphogenesis
Maria E Bustillo, Jessica Douthit, Sergio Astigarraga, Jessica E Treisman
Periods of environmental sensitivity couple larval behavior and development
Denis F. Faerberg, Erin Z. Aprison, Ilya Ruvinsky
Aberrant tissue stiffness impairs neural tube development in Mthfd1l mutant mouse embryos
Yogeshwari S. Ambekar, Carlo Donato Caiaffa, Bogdan Wlodarczyk, Manmohan Singh, Alexander W. Schill, John Steele, Salavat R. Aglyamov, Giuliano Scarcelli, Richard H. Finnell, Kirill V. Larin
Thymus formation in uncharted embryonic territoriesv
Isabel Alcobia, Margarida Gama-Carvalho, Leonor Magalhães, Vitor Proa, Domingos Henrique, Hélia Neves
Ayumi Mure, Yuki Sugiura, Rae Maeda, Kohei Honda, Nozomu Sakurai, Yuuki Takahashi, Masayoshi Watada, Toshihiko Katoh, Aina Gotoh, Yasuhiro Gotoh, Itsuki Taniguchi, Keiji Nakamura, Tetsuya Hayashi, Takane Katayama, Tadashi Uemura, Yukako Hattori
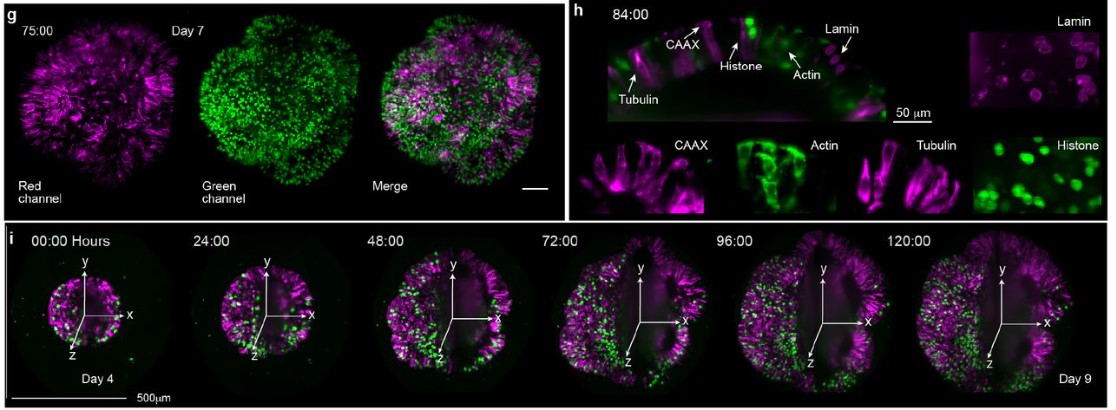
Morphodynamics of human early brain organoid development
Akanksha Jain, Gilles Gut, Fátima Sanchís Calleja, Ryoko Okamoto, Simon Streib, Zhisong He, Fides Zenk, Malgorzata Santel, Makiko Seimiya, René Holtackers, Sophie Martina Johanna Jansen, J. Gray Camp, Barbara Treutlein
Lineage-based scaling of germline intercellular bridges during oogenesis
Umayr Shaikh, Kathleen Sherlock, Julia Wilson, William Gilliland, Lindsay Lewellyn
An atypical basement membrane forms a midline barrier in left-right asymmetric gut development
Cora Demler, John Coates Lawlor, Ronit Yelin, Dhana Llivichuzcha-Loja, Lihi Shaulov, David Kim, Megan Stewart, Frank Lee, Thomas Schultheiss, Natasza Kurpios
Lymphatic vessel development in human embryos
Shoichiro Yamaguchi, Natsuki Minamide, Hiroshi Imai, Tomoaki Ikeda, Masatoshi Watanabe, Kyoko Imanaka-Yoshida, Kazuaki Maruyama
Alternating polarity integrates chemical and mechanical cues to drive tissue morphogenesis
Miriam Osterfield
Xu Zhang, Jerome Avellaneda, Maria Lynn Spletter, Sandra B Lemke, Pierre Mangeol, Bianca H Habermann, Frank Schnorrer
Jyoti Prabha Satta, Qiang Lan, Makoto Mark Taketo, Marja Liisa Mikkola
Simon Schneider, Andjela Kovacevic, Michelle Mayer, Ann-Kristin Dicke, Lena Arévalo, Sophie A. Koser, Jan N. Hansen, Samuel Young, Christoph Brenker, Sabine Kliesch, Dagmar Wachten, Gregor Kirfel, Timo Strünker, Frank Tüttelmann, Hubert Schorle
Xiong Yang, Rong Wan, Zhiwen Liu, Su Feng, Jiaxin Yang, Naihe Jing, Ke Tang
Primate-expressed EPIREGULIN promotes basal progenitor proliferation in the developing neocortex
Paula Cubillos, Nora Ditzer, Annika Kolodziejczyk, Gustav Schwenk, Janine Hoffmann, Theresa M. Schütze, Razvan P. Derihaci, Cahit Birdir, Johannes E. M. Köllner, Andreas Petzold, Mihail Sarov, Ulrich Martin, Katherine R. Long, Pauline Wimberger, Mareike Albert
| Genes & genomes
A. Lardenois, A. Suglia, CL. Moore, B. Evrard, L. Noël, P. Rivaud, A. Besson, M. Toupin, S. Léonard, L. Lesné, I. Coiffec, S. Nef, V. Lavoué, O. Collin, A. Chédotal, S. Mazaud-Guittot, F. Chalmel, AD. Rolland
The TOP-2-condensin II axis silences transcription during germline specification in C. elegans
Mezmur D. Belew, Emilie Chien, Matthew Wong, W. Matthew Michael
Transcriptional control of compartmental boundary positioning during Drosophila wing development
Gustavo Aguilar, Michèle Sickmann, Dimitri Bieli, Gordian Born, Markus Affolter, Martin Müller
Joab Camarena, Sofia E. Luna, Jessica P. Hampton, Kiran R. Majeti, Carsten T. Charlesworth, Eric Soupene, Vivien A. Sheehan, M. Kyle Cromer, Matthew H. Porteus
Defining the cellular complexity of the zebrafish bipotential gonad
Michelle E. Kossack, Lucy Tian, Kealyn Bowie, Jessica S. Plavicki
Michał Brouze, Agnieszka Czarnocka-Cieciura, Olga Gewartowska, Monika Kusio-Kobiałka, Kamil Jachacy, Marcin Szpila, Bartosz Tarkowski, Jakub Gruchota, Paweł Krawczyk, Seweryn Mroczek, Ewa Borsuk, Andrzej Dziembowski
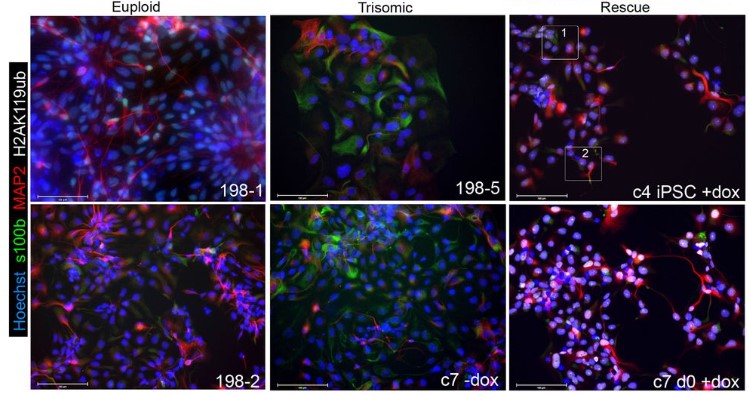
A dynamic in vitro model of Down Syndrome neurogenesis with Trisomy 21 gene dosage correction
Prakhar Bansal, Erin C Banda, Heather R Glatt-Deeley, Christopher Stoddard, Jeremy W Linsley, Neha Arora, Darcy T Ahern, Yuvabharath Kondaveeti, Michael Nicouleau, Miguel Sabariego-Navarro, Mara Dierssen, Steve Finkbeiner, Stefan F Pinter
Dynamics of chromatin accessibility during human first-trimester neurodevelopment
Camiel C.A. Mannens, Lijuan Hu, Peter Lonnerberg, Marijn Schipper, Caleb Reagor, Xiaofei Li, Xiaoling He, Roger A. Barker, Erik Sundstrom, Danielle Posthuma, Sten Linnarsson
Jos Smits, Dulce Lima Cunha, Jieqiong Qu, Nicholas Owen, Lorenz Latta, Nora Szentmary, Berthold Seitz, Lauriane N. Roux, Mariya Moosajee, Daniel Aberdam, Simon J. van Heeringen, Huiqing Zhou
PRDM16 co-operates with LHX2 to shape the human brain
Varun Suresh, Bidisha Bhattacharya, Rami Yair Tshuva, Miri Danan Gotthold, Tsviya Olender, Mahima Bose, Saurabh J. Pradhan, Bruria Ben Zeev, Richard Scott Smith, Shubha Tole, Sanjeev Galande, Corey Harwell, José-Manuel Baizabal, Orly Reiner
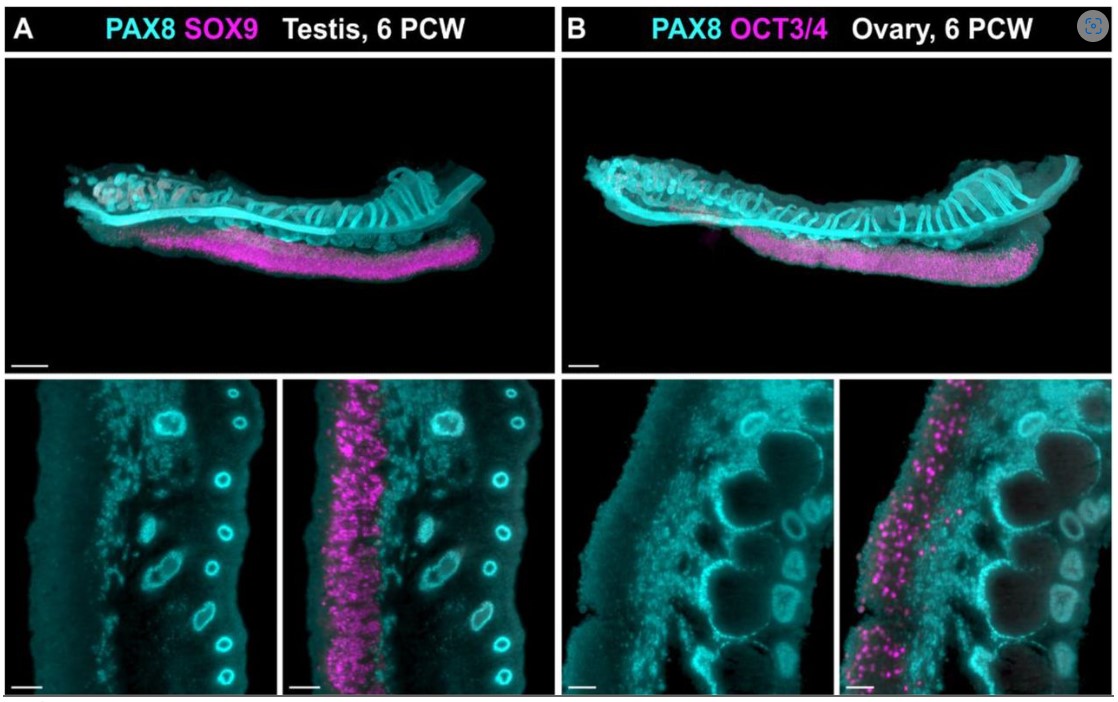
COUP-TFII regulates early bipotential gonad signaling and commitment to ovarian progenitors
Lucas G. A. Ferreira, Marina M. L. Kizys, Gabriel A. C. Gama, Svenja Pachernegg, Gorjana Robevska, Andrew H. Sinclair, Katie L. Ayers, Magnus R. Dias da Silva
Sphingolipid metabolism is spatially regulated in the developing embryo by SOXE genes
Michael L. Piacentino, Aria J. Fasse, Alexis Camacho-Avila, Ilya Grabylnikov, Marianne E. Bronner
Gabriella Martini, Hernan Garcia
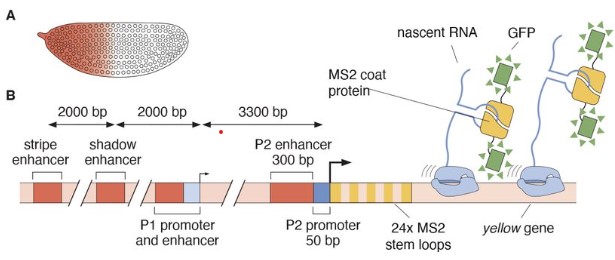
Meng-Ling Wu, Kate Wheeler, Robert Silasi-Mansat, Florea Lupu, Courtney T. Griffin
Female germline expression of OVO transcription factor bridges Drosophila generations
Leif Benner, Savannah Muron, Brian Oliver
Anupama Prakash, Emilie Dion, Antónia Monteiro
FOXL2 interaction with different binding partners regulates the dynamics of ovarian development
Roberta Migale, Michelle Neumann, Richard Mitter, Mahmoud-Reza Rafiee, Sophie Wood, Jessica Olsen, Robin Lovell-Badge
| Stem cells, regeneration & disease modelling
A dynamic in vitro model of Down Syndrome neurogenesis with Trisomy 21 gene dosage correction
Prakhar Bansal, Erin C Banda, Heather R Glatt-Deeley, Christopher Stoddard, Jeremy W Linsley, Neha Arora, Darcy T Ahern, Yuvabharath Kondaveeti, Michael Nicouleau, Miguel Sabariego-Navarro, Mara Dierssen, Steve Finkbeiner, Stefan F Pinter
Ilse Eidhof, Malin Kele, Mansoureh Shahsavani, Benjamin Ulfenborg, Dania Winn, Per Uhlen, Anna Falk
Ece Somuncular, Tsu-Yi Su, Ozge Dumral, Anne-Sofie Johansson, Sidinh Luc
Shlomit Edri, Vardit Rosenthal, Or Ginsburg, Abigail Newman Frisch, Christophe E. Pierreux, Nadav Sharon, Shulamit Levenberg
ECM degradation in the stump region induced by Fgf during functional joint regeneration in frogs
Haruka Matsubara, Takeshi Inoue, Kiyokazu Agata
Alexa Giovannini, Sabrina Piechota, Maria Marchante, Kathryn S Potts, Graham Rockwell, Bruna Paulsen, Alexander D Noblett, Samantha L Estevez, Alexandra B Figueroa, Caroline Aschenberger, Dawn A Kelk, Marcy Forti, Shelby Marcinyshyn, Ferran Barrachina, Klaus Wiemer, Marta Sanchez, Pedro Belchin, Merrick Pierson Smela, Patrick R.J. Fortuna, Pranam Chatterjee, David H McCulloh, Alan Copperman, Daniel Ordonez-Perez, Joshua U Klein, Christian C Kramme
Loss of PAX6 alters the excitatory/inhibitory neuronal ratio in human cerebral organoids
Wai Kit Chan, Danilo Negro, Victoria M Munro, Helen Marshall, Zrinko Kozić, Megan Brown, Mariana Beltran, Neil C Henderson, David J Price, John O Mason
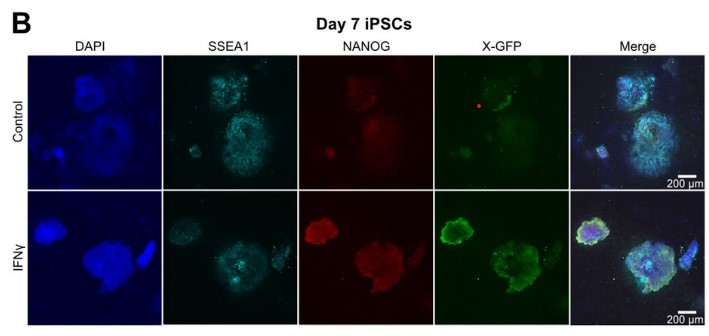
The Interferon γ Pathway Enhances Pluripotency and X-Chromosome Reactivation in iPSC reprogramming
Mercedes Barrero, Anna V. López-Rubio, Aleksey Lazarenkov, Enrique Blanco, Moritz Bauer, Luis G. Palma, Anna Bigas, Luciano Di Croce, José Luis Sardina, Bernhard Payer
Myogenetic Oligodeoxynucleotide Induces Myocardial Differentiation of Murine Pluripotent Stem Cells
Mina Ishioka, Yuma Nihashi, Yoichi Sunagawa, Koji Umezawa, Takeshi Shimosato, Hiroshi Kagami, Tatsuya Morimoto, Tomohide Takaya
Liang-Yu Pang, Steven DeLuca, Haolong Zhu, John M. Urban, Allan C. Spradling
Clair M. Kelley, Nicole O. Glenn, Dafne Gays, Massimo M. Santoro, Wilson K. Clements
A distinct class of hematopoietic stem cells develop from the human yellow bone marrow
Tammy T Nguyen, Zinger Yang Loureiro, Anand Desai, Tiffany DeSouza, Shannon Joyce, Lyne Khair, Amruta Samant, Haley Cirka, Javier Solivan-Rivera, Rachel Ziegler, Michael Brehm, Louis M Messina, Silvia Corvera
Rachel B Gilmore, Dea Gorka, Christopher Stoddard, Justin Cotney, Stormy Chamberlain
Xiaoyu Yang, Ian R. Jones, Poshen B. Chen, Han Yang, Xingjie Ren, Lina Zheng, Bin Li, Yang E. Li, Quan Sun, Jia Wen, Cooper Beaman, Xiekui Cui, Yun Li, Wei Wang, Ming Hu, Bing Ren, Yin Shen
Qixing Zhong, Yao He, Li Teng, Yinqian Zhang, Ting Zhang, Yinbing Zhang, Qinxi Li, Bangcheng Zhao, Daojun Chen, Zhihui Zhong
Connexin 41.8 mediates the correct temporal induction of haematopoietic stem and progenitor cells
Tim Petzold, Masakatsu Watanabe, Julien Y. Bertrand
Investigating the developmental onset of regenerative potential in the annelid Capitella teleta
Alicia A. Boyd, Elaine C. Seaver
Orchestration of pluripotent stem cell genome reactivation during mitotic exit
Silja Placzek, Ludovica Vanzan, David M. Suter
| Plant development
Nutrient levels control root growth responses to high ambient temperature in plants
Sanghwa Lee, Julia Showalter, Ling Zhang, Gaëlle Cassin-Ross, Hatem Rouached, Wolfgang Busch
Does late water deficit induce root growth or senescence in wheat?
Kanwal Shazadi, John T. Christopher, Karine Chenu
NAC1 directs CEP1-CEP3 peptidase expression and modulates root hair growth in Arabidopsis
Diana R. Rodríguez-García, Yossmayer del Carmen Rondón Guerrero, Lucía Ferrero, Andrés Hugo Rossi, Esteban A. Miglietta, Ariel A. Aptekmann, Eliana Marzol, Javier Martínez Pacheco, Mariana Carignani, Victoria Berdion Gabarain, Leonel E. Lopez, Gabriela Díaz Dominguez, Cecilia Borassi, José Juan Sánchez-Serrano, Lin Xu, Alejandro D. Nadra, Enrique Rojo, Federico Ariel, José M. Estevez
BrrRCO, encoding a homeobox protein, is involved in leaf lobe development in Brassica rapa
Pan Li, Tongbing Su, Yudi Wu, Hui Li, Limin Wang, Fenglan Zhang, Shuancang Yu, Zheng Wang
ZmPILS6 is an auxin efflux carrier required for maize root morphogenesis
Craig L. Cowling, Arielle L. Homayouni, Jodi B. Callwood, Maxwell R. McReynolds, Jasper Khor, Haiyan Ke, Melissa A. Draves, Katayoon Dehesh, Justin W. Walley, Lucia C. Strader, Dior R. Kelley
Sukhendu Maity, Rajkumar Guchhait, Kousik Pramanick
Spatiotemporally distinct responses to mechanical forces shape the developing seed of Arabidopsis
Amelie Bauer, Camille Bied, Adrien Delattre, Gwyneth Ingram, John F Golz, Benoit Landrein
Chitosan stimulates root hair callose deposition and inhibits root hair growth
Matēj Drs, Samuel Haluška, Eliška Škrabálková, Pavel Krupař, Andrea Potocká, Lucie Brejšková, Karel Muller, Natalia Serrano, Aline Voxeur, Samantha Vernhettes, Jitka Ortmannová, George Caldarescu, Matyas Fendrych, Martin Potocký, Viktor Žárský, Tamara Pečenková
A pectin-binding peptide with a structural and signaling role in the assembly of the plant cell wall
Sebastjen Schoenaers, Hyun Kyung Lee, Martine Gonneau, Elvina Faucher, Thomas Levasseur, Elodie Akary, Naomi Claeijs, Steven Moussu, Caroline Broyart, Daria Balcerowicz, Hamada AbdElgawad, Andrea Bassi, Daniel Santa Cruz Damineli, Alex Costa, Jose A Feijo, Celine Moreau, Estelle Bonnin, Bernard Cathala, Julia Santiago, Herman Hofte, Kris Vissenberg
Ancy E.J. Chandran, Aliza Finkler, Tom Aharon Hait, Yvonne Kiere, Sivan David, Metsada Pasmanik-Chor, Doron Shkolnik
Samuel Knosp, Lucie Kriegshauser, Kanade Tatsumi, Ludivine Malherbe, Gertrud Wiedemann, Bénédicte Bakan, Takayuki Kohchi, Ralf Reski, Hugues Renault
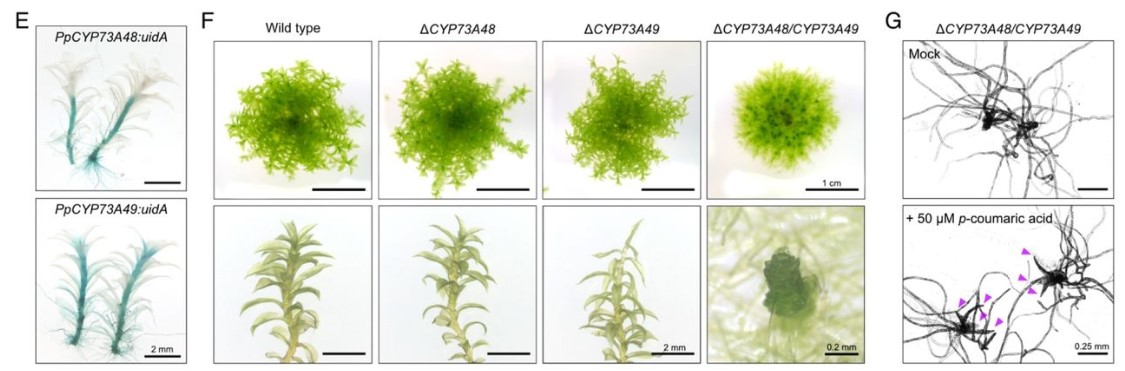
Simon Amiard, Léa Feit, Lauriane Simon, Samuel Le Goff, Loriane Loizeau, Léa Wolff, Falk Butter, Clara Bourbousse, Fredy Barneche, Christophe Tatout, Aline. V. Probst
The Auxin Response Factor ARF27 is required for maize root morphogenesis
Linkan Dash, Maxwell R. McReynolds, Melissa A. Draves, Rajdeep S. Khangura, Rebekah L. Muench, Jasper Khor, Jodi B. Callwood, Craig L. Cowling, Ludvin Mejia, Michelle G. Lang, Brian P. Dilkes, Justin W. Walley, Dior R. Kelley
Overexpression of tomato SlBBX16 and SlBBX17 impacts fruit development and GA biosynthesis
Valentina Dusi, Federica Pennisi, Daniela Fortini, Alejandro Atarès, Stephan Wenkel, Barbara Molesini, Tiziana Pandolfini
Growth directions and stiffness across cell layers determine whether tissues stay smooth or buckle
Avilash S. Yadav, Lilan Hong, Patrick M. Klees, Annamaria Kiss, Xi He, Iselle M. Barrios, Michelle Heeney, Anabella Maria D. Galang, Richard S. Smith, Arezki Boudaoud, Adrienne H.K. Roeder
Water fluxes contribute to growth patterning in shoot meristems
Juan Alonso-Serra, Ibrahim Cheddadi, Annamaria Kiss, Guillaume Cerutti, Claire Lionnet, Christophe Godin, Olivier Hamant
Lisa Oskam, Basten L. Snoek, Chrysoula K. Pantazopoulou, Hans van Veen, Sanne E. A. Matton, Rens Dijkhuizen, Ronald Pierik
Cell Fate Programming by Transcription Factors and Epigenetic Machinery in Stomatal Development
Ao, Andrea Mair, Juliana L. Matos, Macy Vollbrecht, Shou-Ling Xu, Dominique C. Bergmann
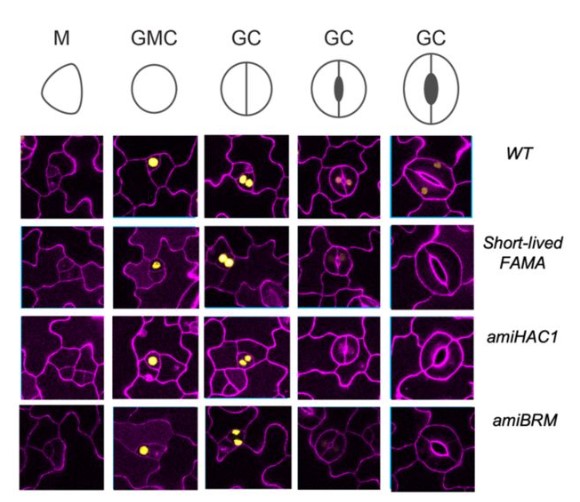
| Evo-devo
Stacy V Nguyen, Rachel S Lee, Emma Mohlmann, Gabriella Petrullo, John Blythe, Isabella Ranieri, Sarah McMenamin
Shiho Takahashi-Kariyazono, Akira Iguchi, Yohey Terai
Clearwing butterflies challenge the thermal melanism hypothesis
Violaine Ossola, Fabien Pottier, Charline Pinna, Katia Bougiouri, Aurélie Tournié, Anne Michelin, Christine Andraud, Doris Gomez, Marianne Elias
A mathematical framework for evo-devo dynamics
Mauricio González-Forero
Conserved switch genes regulate a novel cannibalistic morph after whole genome duplication
Sara Wighard, Ralf J. Sommer, Hanh Witte
Nathan D. Harry, Christina Zakas
Amber M. Ridgway, Emily Hood, Javier Figueras Jimenez, Maria D. S. Nunes, Alistair P. McGregor
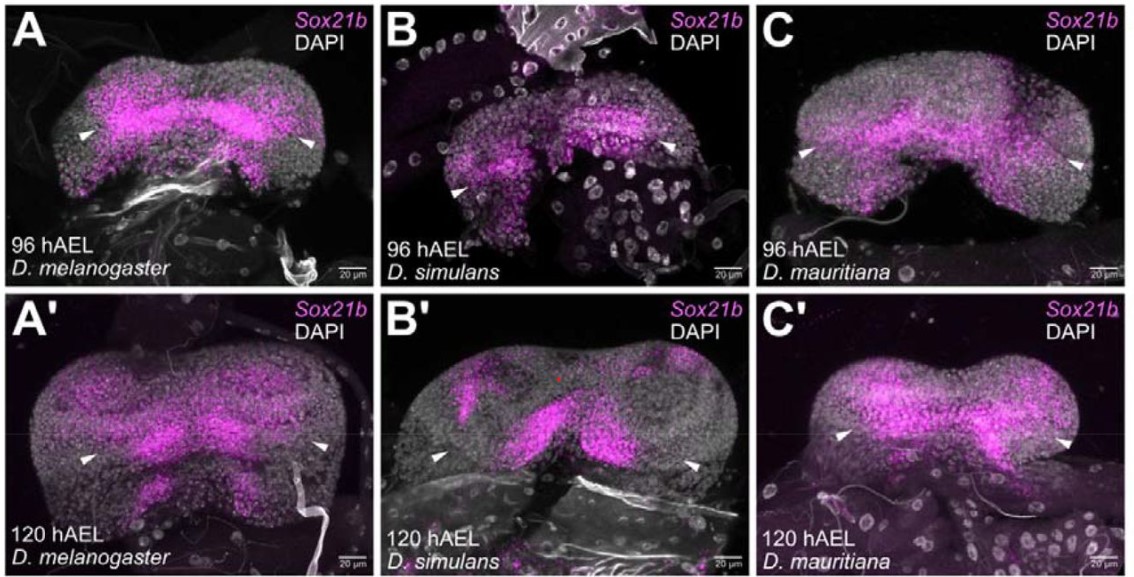
Evo-devo dynamics of hominin brain size
Mauricio González-Forero
Evolution of parent-of-origin effects on placental gene expression in house mice
Fernando Rodriguez-Caro, Emily C. Moore, Jeffrey M. Good
Cell Biology
David Bolumar, Javier Moncayo-Arlandi, Javier Gonzalez-Fernandez, Ana Ochando, Inmaculada Moreno, Ana Monteagudo-Sanchez, Carlos Marin, Antonio Diez, Paula Fabra, Miguel Ángel Checa, Juan José Espinos, David K. Gardner, Carlos Simon, Felipe Vilella
Robo1 loss has pleiotropic effects on postnatal development and survival
Nicole A. Kirk, Ye Eun Hwang, Seul Gi Lee, Kee-Beom Kim, Kwon-Sik Park
Actomyosin-mediated apical constriction promotes physiological germ cell death in C. elegans
Tea Kohlbrenner, Simon Berger, Tinri Aegerter-Wilmsen, Ana Laranjeira, Andrew deMello, Alex Hajnal
Isabel Tundidor, Marta Seijo-Vila, Sandra Blasco-Benito, María Rubert-Hernández, Gema Moreno-Bueno, Laura Bindila, Rubén Fernández de la Rosa, Manuel Guzmán, Cristina Sánchez, Eduardo Pérez-Gómez
Purine biosynthesis pathways are required for myogenesis in Xenopus laevis
Maëlle Duperray, Elodie Henriet, Christelle Saint-Marc, Eric Boué-Grabot, Bertrand Daignan-Fornier, Karine Massé, Benoît Pinson
Cyclin B3 is a dominant fast-acting cyclin that drives rapid early embryonic mitoses
Pablo Lara-Gonzalez, Smriti Variyar, Jacqueline Budrewicz, Aleesa Schlientz, Neha Varshney, Andrew Bellaart, Shabnam Moghareh, Anh Cao Ngoc Nguyen, Karen Oegema, Arshad Desai
Neuronal IL-17 controls C. elegans developmental diapause through CEP-1/p53
Abhishiktha Godthi, Sehee Min, Das Srijit, Johnny Cruz-Corchado, Andrew Deonarine, Kara Misel-Wuchter, Priya D. Issuree, Veena Prahlad
Reorganization of Septin structures regulates early myogenesis
Vladimir Ugorets, Paul-Lennard Mendez, Dmitrii Zagrebin, Giulia Russo, Yannic Kerkhoff, Tim Herpelinck, Georgios Kotsaris, Jerome Jatzlau, Sigmar Stricker, Petra Knaus
Modelling
A model-based assessment of adaptation in embryonic life histories of annual killifish
Tom JM Van Dooren
How a reaction-diffusion signal can control spinal cord regeneration in axolotls: A modelling study
Valeria Caliaro, Diane Peurichard, Osvaldo Chara
Spectral decomposition unlocks ascidian morphogenesis
Joel Dokmegang, Emmanuel Faure, Patrick Lemaire, Edwin Munro, Madhav Mani
Manica Balant, Teresa Garnatje, Daniel Vitales, Oriane Hidalgo, Daniel H. Chitwood
Causal models of human growth and their estimation using temporally-sparse data
John A. Bunce, Catalina I. Fernández, Caissa Revilla-Minaya
Morphogen gradients can convey position and time in growing tissues
Roman Vetter, Dagmar Iber
Pierre Galipot
Tools & Resources
Angelica Miglioli, Marion Tredez, Manon Boosten, Camille Sant, Joao E. Carvalho, Philippe Dru, Laura Canesi, Michael Schubert, Remi Dumollard
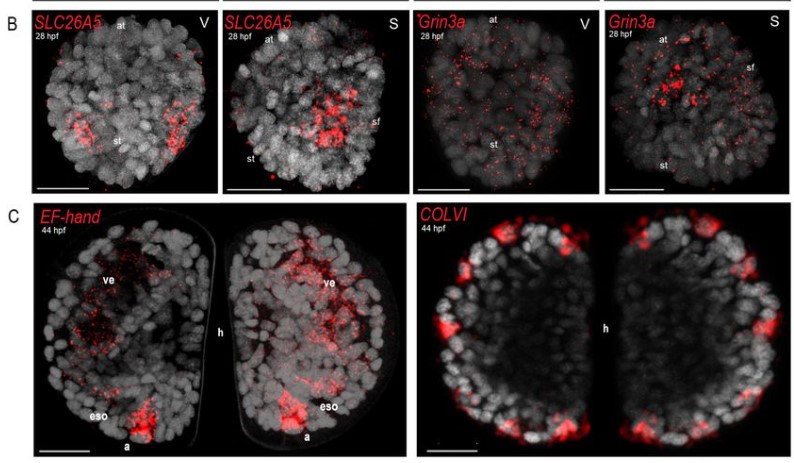
Amanda A. G. Ferreira, Claude Desplan
CONDITIONALLY MUTANT ANIMAL MODEL FOR INVESTIGATING THE INVASIVE TROPHOBLAST CELL LINEAGE
Khursheed Iqbal, Brandon Nixon, Benjamin Crnkovich, Esteban M. Dominguez, Ayelen Moreno-Irusta, Regan L. Scott, Ha T.H. Vu, Geetu Tuteja, Jay L. Vivian, Michael J. Soares
Screening for variable drug responses using human iPSC cohorts
Melpomeni Platani, Hao Jiang, Lindsay Davidson, Santosh Hariharan, Regis Doyonnas, Angus I. Lamond, Jason R. Swedlow
Taro Ichimura, Taishi Kakizuka, YuKi Sato, Keiko Itano, Kaoru Seiriki, Hitoshi Hashimoto, Hiroya Itoga, Shuichi Onami, Takeharu Nagai
Clonal and Scalable Endothelial Progenitor Cell Lines from Human Pluripotent Stem Cells
Jieun Lee, Hal Sternberg, Paola A. Bignone, James Murai, Nafees N. Malik, Michael D. West, Dana Larocca
Early spiral arteriole remodeling in the uterine-placental interface: a rat model
Sarah J. Bacon, Yuxi Zhu, Priyanjali Ghosh
Automated staging of zebrafish embryos with deep learning
Rebecca A. Jones, Matthew J. Renshaw, Danelle Devenport, David J. Barry
A simplified and rapid in situ hybridization protocol for planarians
Andrew J. Gaetano, Ryan S. King
Madeleine N. Hewitt, Iván A. Cruz, David W. Raible
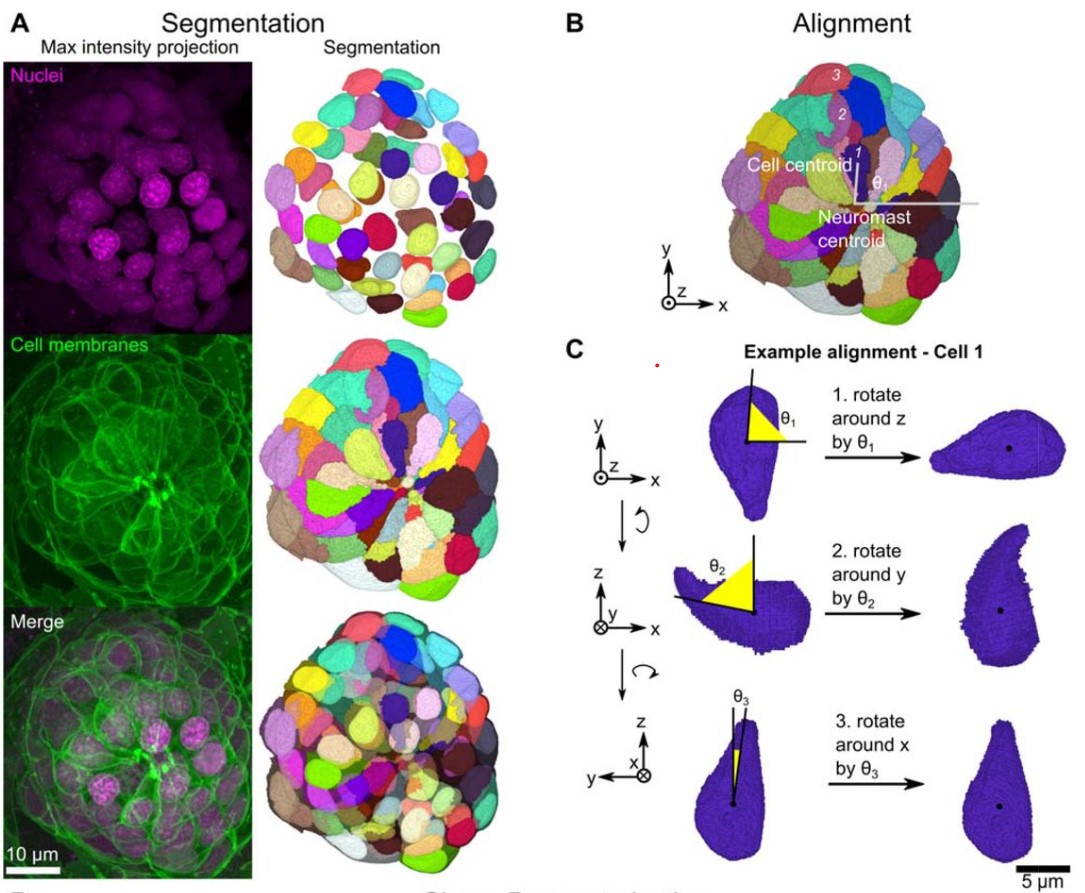
Jesús A. Andrés-San Román, Carmen Gordillo-Vázquez, Daniel Franco-Barranco, Laura Morato, Cecilia H. Fernández-Espartero, Gabriel Baonza, Antonio Tagua, Pablo Vicente-Munuera, Ana M. Palacios, Maria P. Gavilan, Fernando Martín-Belmonte, Valentina Annese, Pedro Gómez-Gálvez, Ignacio Arganda-Carreras, Luis M. Escudero
Research practice & education
Is N-Hacking Ever OK? A simulation-based inquiry
Pamela Reinagel
ChatGPT applications in Academic Research: A Review of Benefits, Concerns, and Recommendations
Adhari AlZaabi, Amira ALAmri, Halima Albalushi, Ruqaya Aljabri, AbdulRahman AalAbdulsalam
Gender differences in submission behavior exacerbate publication disparities in elite journals
Isabel Basson, Chaoqun Ni, Giovanna Badia, Nathalie Tufenkji, Cassidy R. Sugimoto, Vincent Larivière
ChatGPT identifies gender disparities in scientific peer review
Jeroen P. H. Verharen
Analytical code sharing practices in biomedical research
Nitesh Kumar Sharma, Ram Ayyala, Dhrithi Deshpande, Yesha M Patel, Viorel Munteanu, Dumitru Ciorba, Andrada Fiscutean, Mohammad Vahed, Aditya Sarkar, Ruiwei Guo, Andrew Moore, Nicholas Darci-Maher, Nicole A Nogoy, Malak S. Abedalthagafi, Serghei Mangul
Lisa DaVia Rubenstein, Kelsey A. Woodruff, April M. Taylor, James B. Olesen, Philip J. Smaldino, Eric M. Rubenstein
Sarvenaz Sarabipour, Steven J Burgess, Natalie M Niemi, Christopher T Smith, Alexandre W Bisson Filho, Ahmed Ibrahim, Kelly Clark
Graduate student mentorship as a target for diversifying the biological sciences
Reena Debray, Emily Dewald-Wang, Katherine Ennis
Tianqi Lu, Zafar I. Bashir, Alessia Dalceggio, Caroline M. McKinnon, Lydia Miles, Amy Mosley, Bronwen R. Burton, Alice Robson


 (No Ratings Yet)
(No Ratings Yet)

 (2 votes)
(2 votes)